The Community Structure and Diversity Characteristics of Rhizosphere Bacteria and Endophytic Bacteria in Phyllostachys edulis under Annual Growth Characteristics (On and Off Years)
-
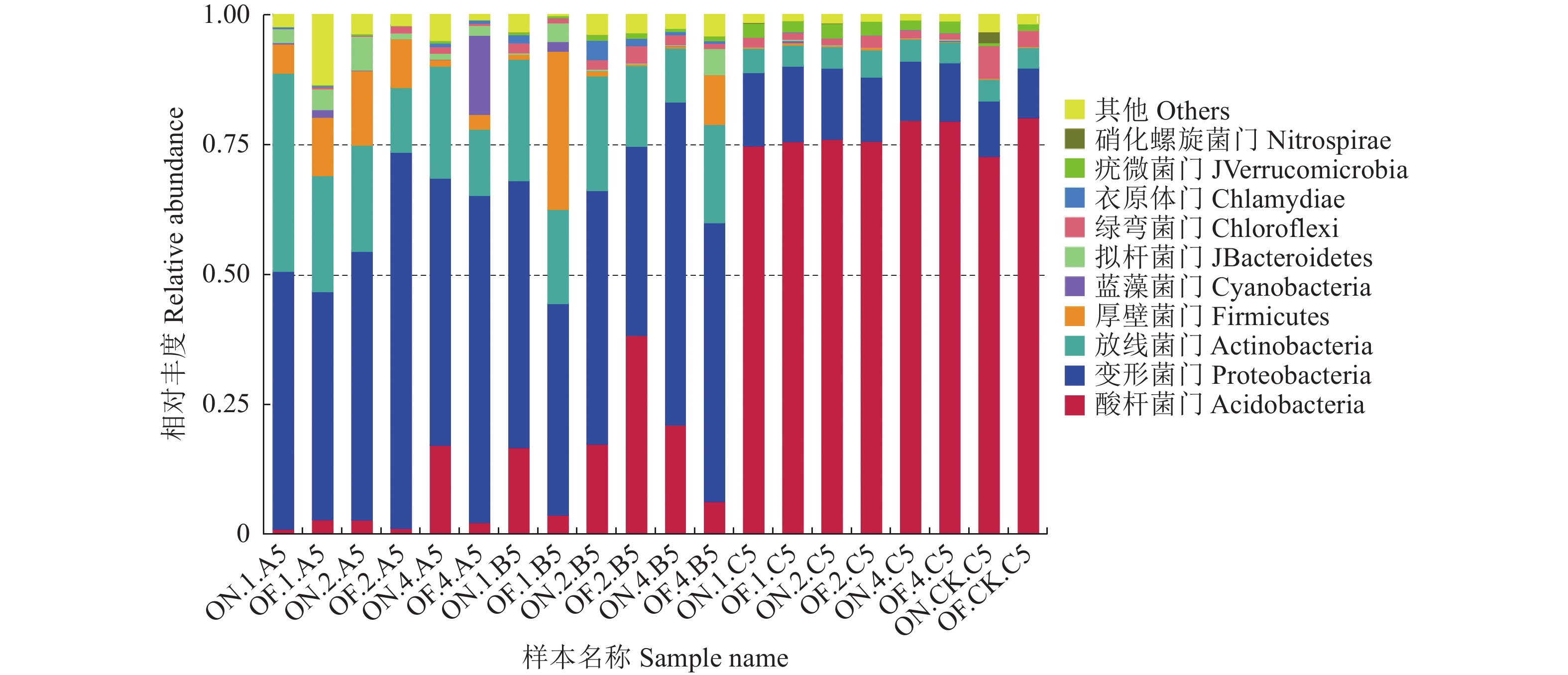
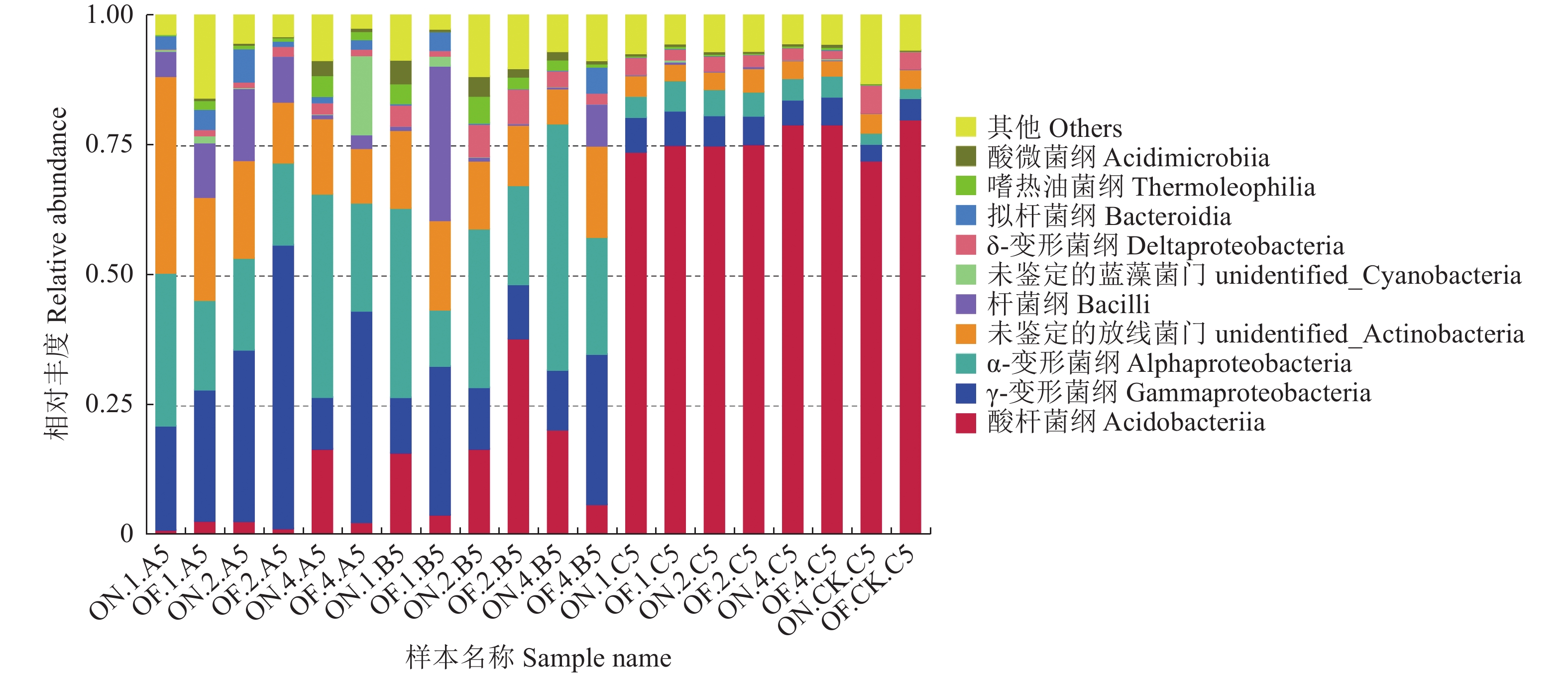
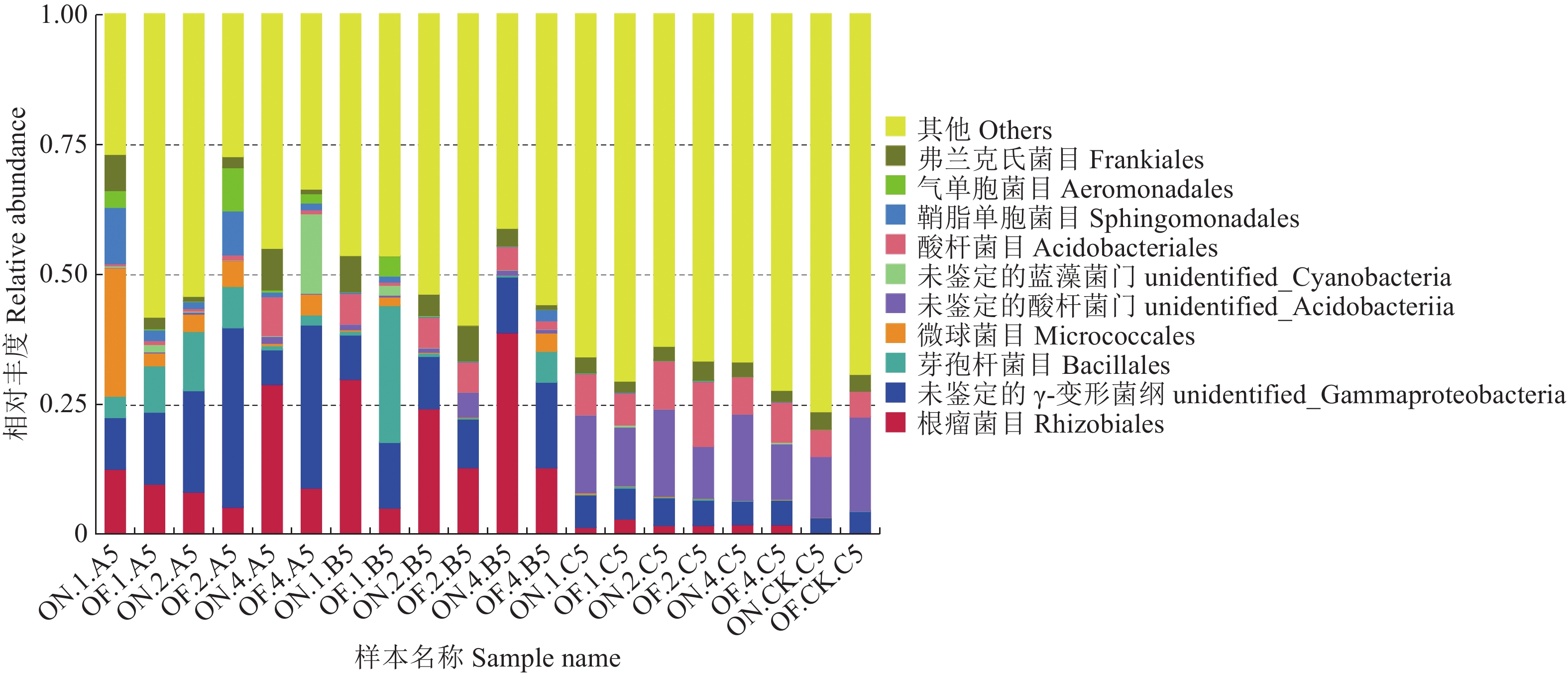
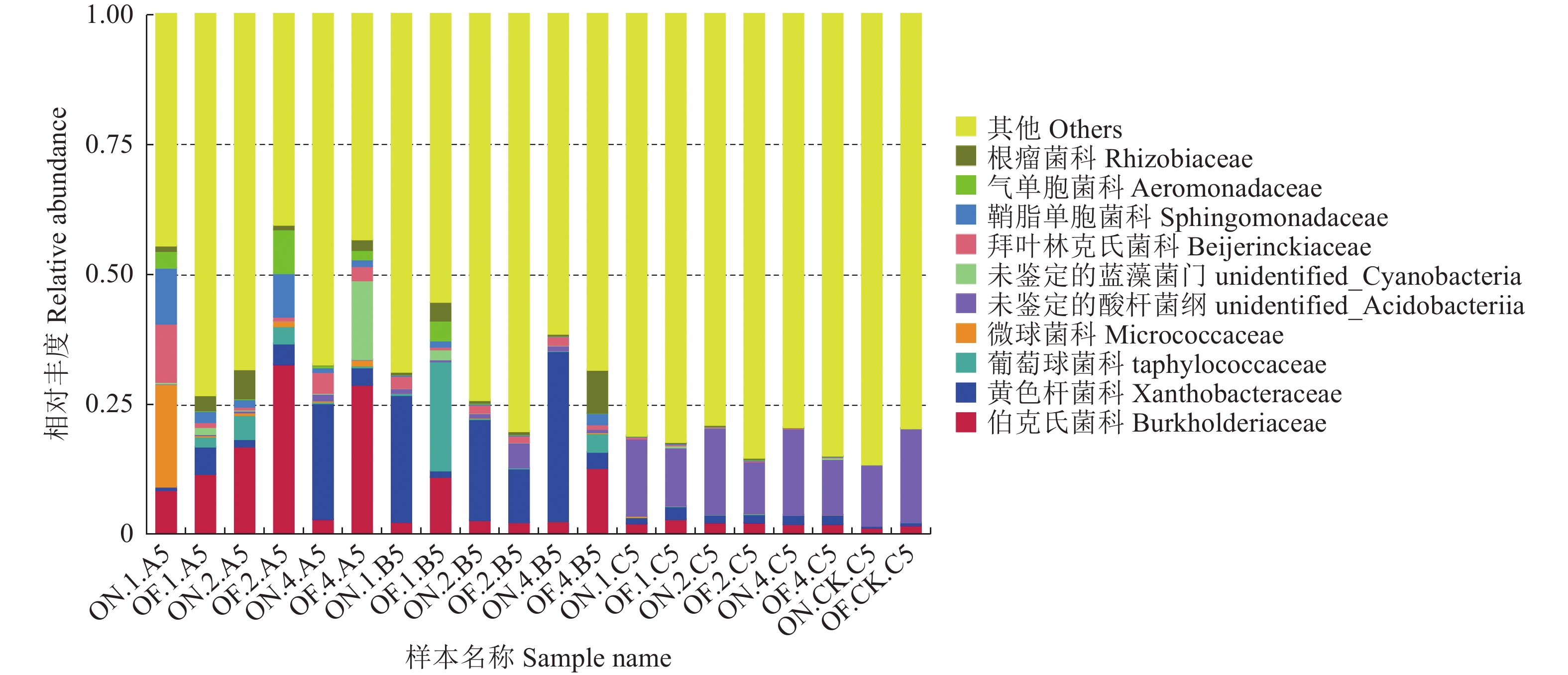
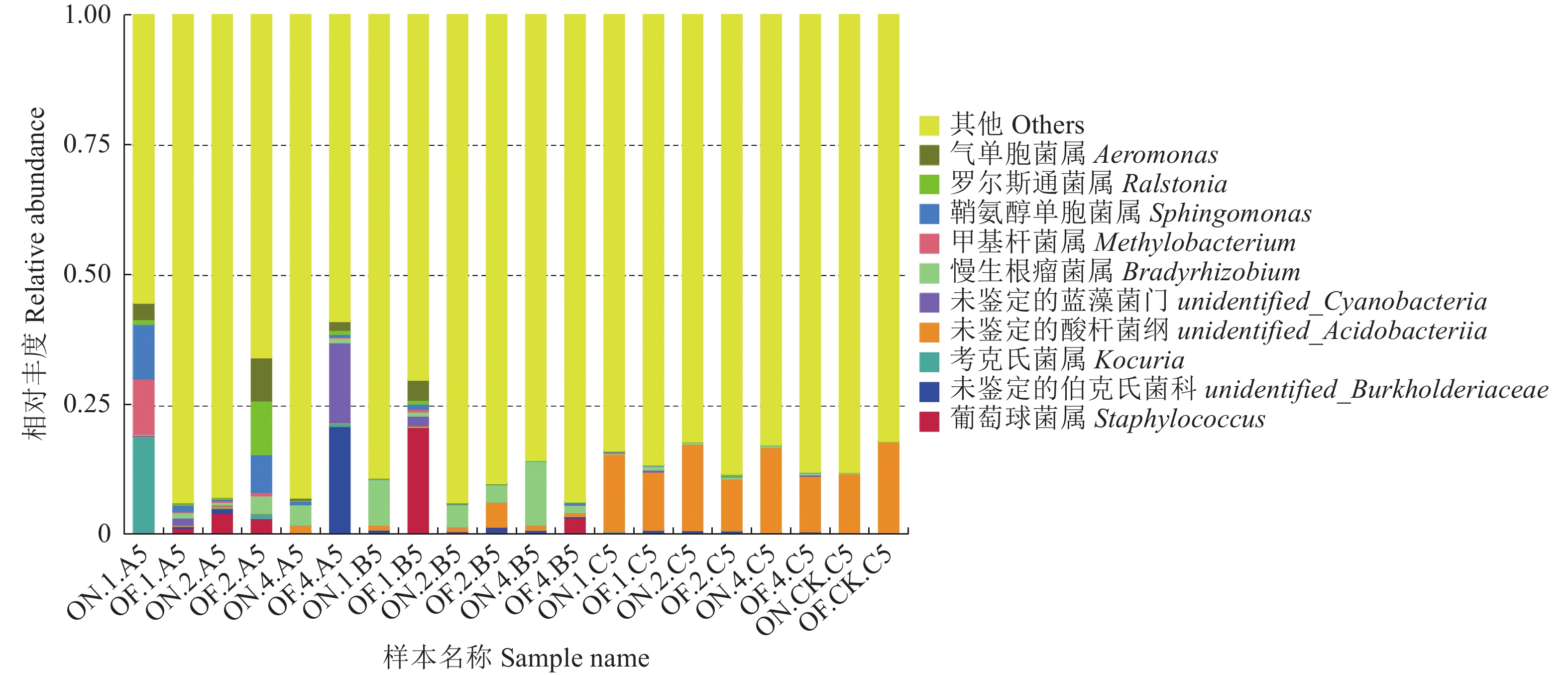
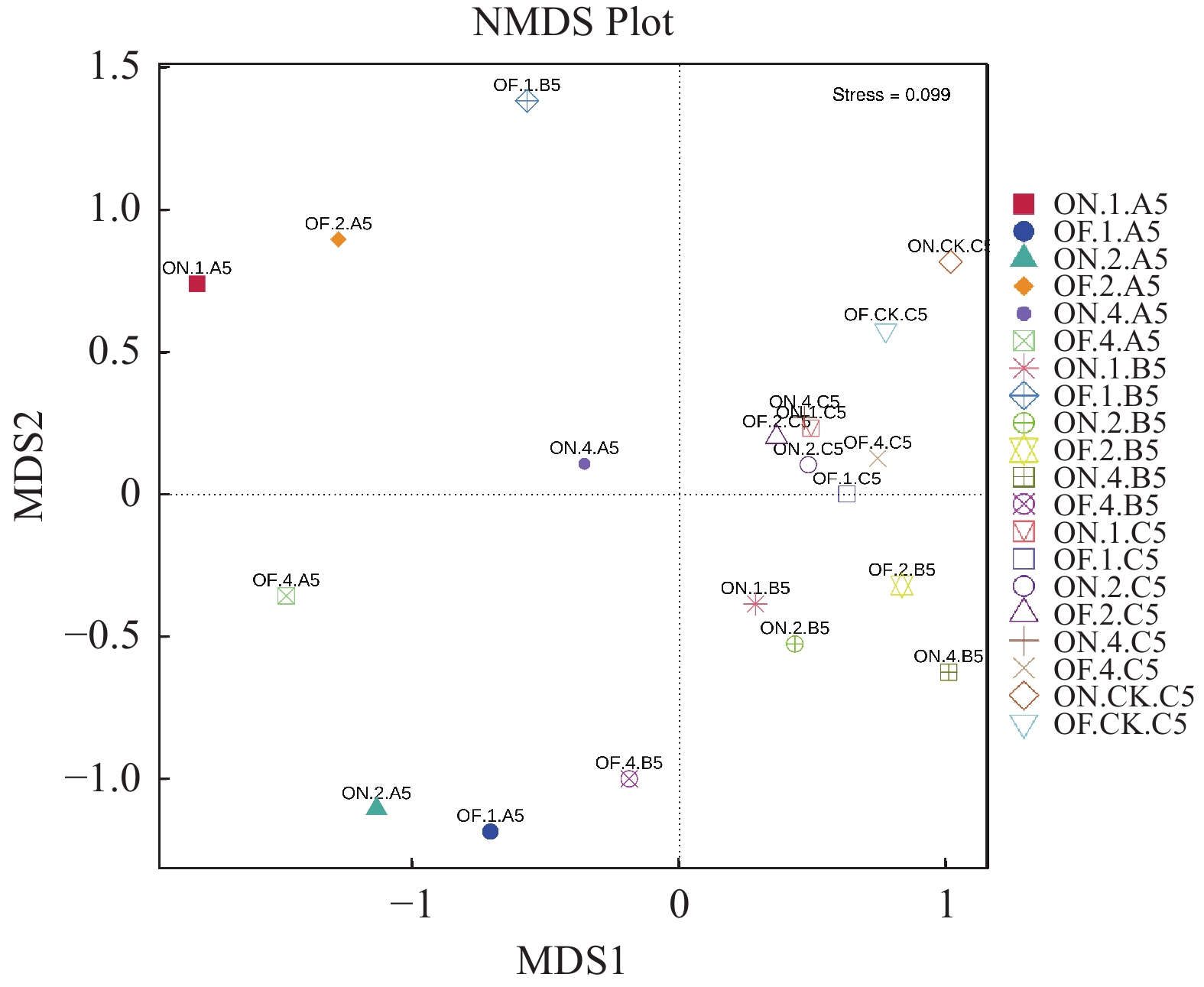
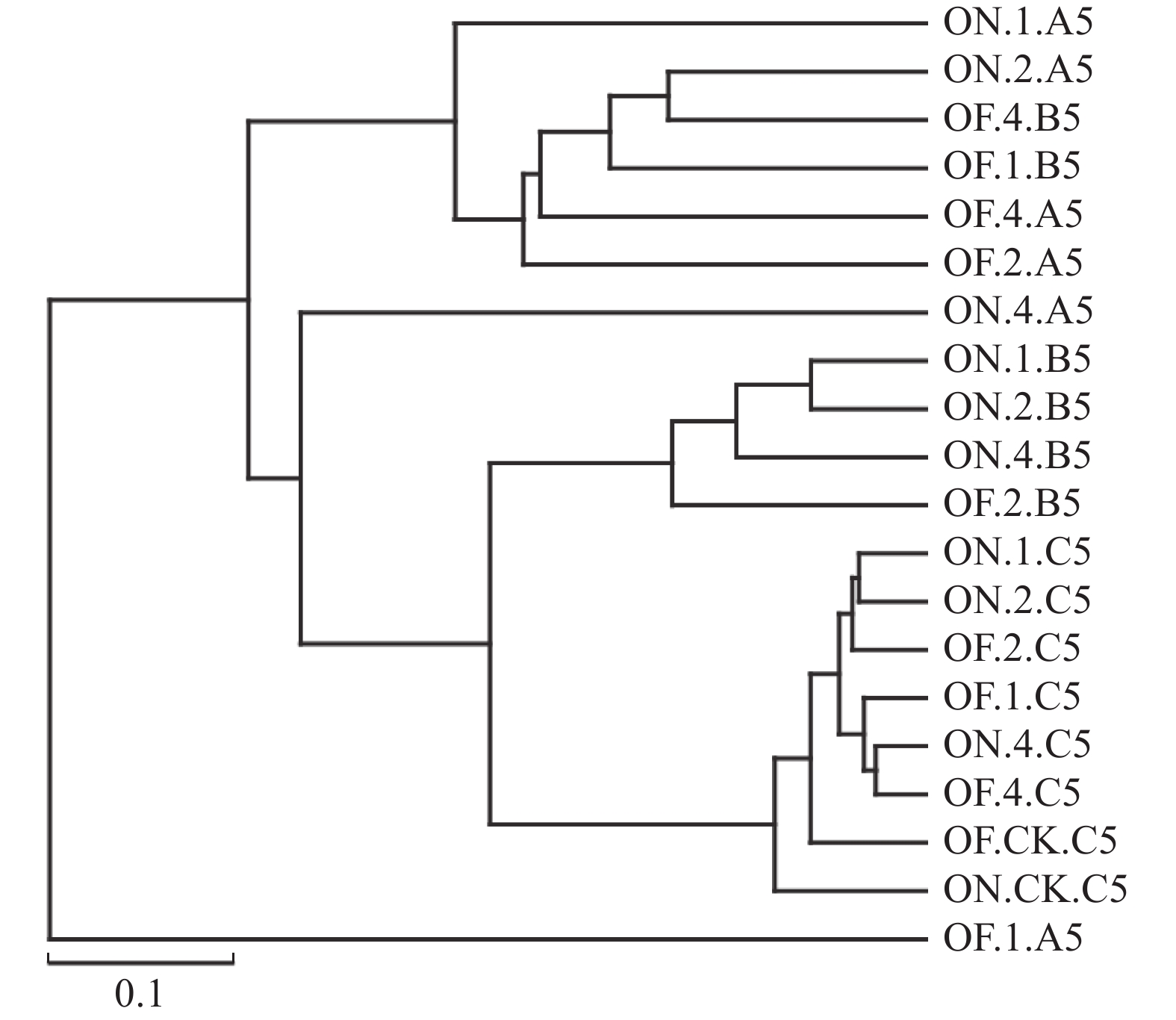
摘要:目的 研究大小年毛竹林毛竹根际细菌和内生细菌群落多样性及其结构差异。方法 采集I度、II度和IV度的大年和小年毛竹林中毛竹的竹鞭、鞭根、根际土壤和林间土壤,提取样本基因组DNA,利用Illumina高通量测序技术分析毛竹根际细菌和内生细菌群落结构多样性。结果 各组样本总共鉴定出31个门、49个纲、108个目、212个科、472个属细菌。从优势菌群及丰度来看,大年竹鞭和鞭根的优势菌纲为α-变形菌纲,优势菌目为根瘤菌目;小年竹鞭和鞭根的优势菌纲为γ-变形菌纲,优势菌目为芽孢杆菌目。在门水平上,大年竹鞭样本放线菌门的丰度高于小年竹鞭样本,大年毛竹鞭根酸杆菌门和变形菌门的丰度大于小年毛竹鞭根样本,厚壁菌门和拟杆菌门的丰富度小于小年毛竹鞭根样本。在纲和目水平上,大年竹鞭和鞭根样本与小年样本相比较,主要优势菌群为弗兰克氏菌目和α-变形菌纲下属的根瘤菌目。在科水平上,大年毛竹鞭根样本在黄杆菌科的丰度都大于小年样本。在属水平上,大年毛竹鞭根样本在慢生根瘤菌属的丰度大于小年毛竹鞭根样本,而大年毛竹竹鞭和鞭根样本细菌在伯克氏菌科的丰度都低于小年毛竹竹鞭和鞭根样本。从多样性来看,大小年毛竹根际土壤在各水平的细菌群落组成上差异不大,但根际细菌的多样性和丰度高于林间土壤。结论 毛竹的竹龄及大小年更替对根际细菌群落多样性的影响不大,根际细菌群落具有更高的多样性。大小年毛竹竹鞭和鞭根内生细菌在主要类群上有明显的不同。Abstract:Objective To study the diversity and structural differences of rhizosphere bacteria and endophytic bacterial communities in Phyllostachys edulis forests during on and off years.Methods Samples of rhizomes and rhizomes roots as well as rhizosphere soils of degree I, degree II and degree IV P. edulis and non-rhizosphere soils were collected in the on and off years. Genomic DNA was extracted from samples, and Illumina high-throughput sequencing technology was used to analyze the diversity of rhizosphere bacteria and endophytic bacterial communities in P. edulis.Results A total of 31 phyla, 49 classes, 108 orders, 212 families, and 472 genera were identified. The dominant phyla in the on-year P. edulis rhizomes and rhizomes roots were α-Amastigotes, and the dominant order Rhizobia. In the off-year specimens, the dominant phylum was γ-Amastigotes, and the dominant order Bacillariophyceae. At phylum level, the abundance of Actinobacteria was higher in the on-year than in off-year rhizomes; and those of Acidobacteria and Methylobacteria were greater in the on-year than in the off-year rhizomes roots; and those of Firmicutes and Bacteroidetes were less in the on-year than in the off-year rhizomes roots. At class and order levels, the dominants included Frankiaceae and Rhizobia of α-Amastigotes in the on-year rhizomes and rhizomes roots as compared with the off-year samples. At the family level, the abundance of Flavobacteriaceae in the rhizomes root of the on-year was greater than that in the off-year samples. At the genus level, the abundance of Bradyrhizobium in the rhizomes root of on-year was greater than that in off-year. However, the abundance of Burkholderiaceae was lower of on-year than off-year. The rhizosphere soil at the forest in either on or off years did not differ significantly on bacterial diversity, but it was higher on the diversity and richness than the non-rhizosphere soil.Conclusion The rhizosphere bacterial community at a P. edulis forest appeared to be more diverse than the non-rhizosphere, although the diversity was not significantly altered between the years of on and off on the P. edulis growth. The dominant bacteria in the rhizomes and rhizomes roots of the plants differed significantly during the on and off years.
-
Keywords:
- Phyllostachys edulis forest /
- on and off year /
- bacterial community /
- diversity
-
0. 引言
【研究意义】猪轮状病毒(porcine rotavirus, PoRV)属于呼肠孤病毒科(Reoviridae)轮状病毒属(Rotaviruses),基因组由11个双链RNA片段组成,轮状病毒根据VP6基因的核苷酸同源性分为10个群(RVA~RVJ),其中以RVA感染为主[1]。猪A群轮状病毒(porcine rotavirus group A, PoRV A)是引起猪群病毒性腹泻的重要病原体之一[2],主要通过粪口传播,感染的猪群表现为水样稀粪、呕吐等症状,严重者脱水死亡,持续发病可导致猪群严重消瘦[3],解剖病猪可以观察到肠管变薄、肠系淋巴结肿大等病变[4]。猪轮状病毒可感染不同阶段的猪群,并且每只猪可经历一次或多次轮状病毒感染的情况。哺乳仔猪对PoRV病毒特别易感,尤其是不足一周龄的仔猪,不仅最易感染,而且一旦感染,病情也最为严重,其病死率可达100%;相比之下,成年猪感染PoRV时往往表现为无症状的隐性感染[5]。目前,由于PoRV复杂的基因型[1],市场上缺少针对PoRV的特效药,因此建立快速、灵敏、高效的PoRV检测手段,对该病的防治具有重要意义。【前人研究进展】PoRV检测方法主要有病毒分离、免疫荧光、普通PCR等技术。病毒分离技术在研究和防疫检测中发挥着重要作用[6],但是操作技术复杂、消耗成本高、耗时;免疫荧光技术,利用荧光标记的抗体来检测和定位细胞或组织中的特定抗原[7],荧光标记的抗体在长时间的光照下可能会发生荧光淬灭,导致信号减弱,结果判定不准确;普通PCR方法处理样本时间长,结果可能出现假阳性[8]。这些传统方法操作复杂、耗时长等缺点,在检测上具有一定的局限性[9]。【本研究切入点】猪A群轮状病毒的VP6基因适用于多种猪A群轮状病毒亚型的检测[10]。近年来,轮状病毒的高重组率引发了VP6基因在遗传进化上的变化,尽管VP6基因通常被认为高度保守,但已有研究证实该基因也存在基因重组现象[11]。本研究参考了近年来新出现的毒株以及经典毒株的VP6基因片段保守区,设计的TaqMan荧光定量RT-PCR方法既保留了对经典毒株的检测能力,也能有效应对较新的毒株。【拟解决的关键问题】本研究基于猪A群轮状病毒VP6基因,建立适用于PoRV检测及其流行病学调查的TaqMan实时荧光定量PCR检测方法。
1. 材料与方法
1.1 核酸和临床样品
猪轮状病毒(PoRV)、猪流行性腹泻病毒(porcine epidemic diarrhea virus, PEDV)、猪德尔塔冠状病毒(porcine deltacorona virus, PDCoV)、猪传染性胃肠炎病毒(transmissible gastroenteritis of swine, TGEV)核酸,由福建省农业科学院畜牧兽医所猪病研究室保存。疑似PoRV感染的临床样品151份,包括肠道和粪便样品,来源于福建省福州、三明、龙岩、泉州、南平、宁德等地的规模化猪场,−80 ℃保存。
1.2 试剂
SimplyP病毒RNA提取试剂盒(BSC56S1)购自博日科技股份有限公司(杭州);HiScript II One Step RT-PCR Kit试剂盒(P612)购自生物科技股份有限公司(南京);One Step RT-qPCR Probe Kit(BBI,B639278)购自生工生物工程有限公司(上海)。
1.3 引物设计
根据GenBank 中已有的PoRV A VP6基因的序列(登录号MT025937.1、OP978242.1、PP566178.1),并用NCBI BLAST比对选取保守区域,利用Oligo 7和Primer 5设计引物(PoRV-F3、PoRV-R3)及探针(PoRV-Probe)用于TaqMan实时荧光定量RT-PCR体系的建立(表1)。参考Tao等[12]所描述的方法合成针对PoRV VP6基因的特异性引物(PoRV-F2、PoRV-R2),用于逆转录聚合酶链反应(RT-PCR)检测。引物和探针由尚亚生物技术有限公司(福州)合成。
表 1 PoRV引物和探针Table 1. PoRV primers and probes引物/探针
Primer/Probe引物序列(5′-3′)
Sequence(5′-3′)产物大小
Product size/bp用途
UsagePoRV-F1 GGCTTTTAAACGAAGTCTTC 1209 RT-PCR PoRV-R1 GGTCACATCCTCTCACT PoRV-F2 GGCTTTTAAACGAAGTCTTC 598 RT-PCR PoRV-R2 CCAGCTACYTGAATTTCTGA PoRV-F3 ATTAAGTGAGGACTAGGCTAA 114 RT-qPCR PoRV-R3 ACTCTACGTAGCGAGTATGA PoRV-Probe FAM-ATGTAGCTATGTCAAGTCAATCAGA-TAMRA RT-qPCR 1.4 PoRV A阳性质粒构建
利用病毒核酸提取试剂盒(BSC56S1)提取病毒总RNA,使用诺维赞一步法试剂盒(P612)扩增PoRV A VP6基因片段。扩增程序:50 ℃ 30 min,94 ℃ 3 min;94 ℃ 15 s,54 ℃ 30 s,72 ℃ 30 s(35个循环);72 ℃ 7 min。扩增体系:2×One Step Mix 25.0 μL,模板 5.0 μL,One Step Enzyme Mix 2.5 μL,引物PoRV- F1、PoRV- R1(10 μmol·L−1)各2.0 μL,RNase-free ddH2O 添加至 50.0 μL。经琼脂糖凝胶电泳及凝胶成像系统分析,回收纯化PCR产物,连接至pUC57载体。质粒转化至DH5α感受态细胞中,并在LB液体培养基中以37 ℃、180 r·min−1的条件培养,提取转化后感受态细胞中的重组质粒。含有VP6靶向基因序列的阳性质粒通过琼脂糖凝胶电泳与凝胶成像系统分析,并送至尚亚生物技术有限公司(福州)测序证实是否成功构建。
1.5 反应体系的优化
通过测定最低Ct值和最高荧光强度判定优化结果。将引物对(PoRV- F3、PoRV- R3)与探针(PoRV-Probe)分别稀释至1、3、6、9、10 μmol·L−1,以筛选出最优的引物浓度、探针浓度。反应体系根据试剂说明书推荐的一步法反转录荧光定量探针试剂盒20 μL体系,以2.7×104 copies·μL−1拷贝数的阳性质粒为模板,并设置阴性对照。
1.6 标准曲线的绘制
将标准品阳性质粒按10倍倍比稀释,将稀释好的(107~103)浓度质粒作为模板。标准曲线的绘制与计算参考陈秋勇等[13]的方法。
1.7 特异性试验
根据优化后的反应体系进行实时荧光定量RT-PCR扩增,以PEDV、TGEV、PDCoV、PoRV A的RNA为模板,同时设置阴性对照,以评价该检测方法的特异性。
1.8 敏感性试验
将标准品阳性质粒按10倍倍比稀释,将稀释好的质粒(浓度为108~101)为模板,同时加入阴性对照,分别进行实时荧光定量RT-PCR反应和RT-PCR反应,比较两种方法能够检测到的最低浓度,评估其灵敏度。
1.9 重复性试验
取重组质粒浓度为 2.7×105 copies·μL−1、2.7×104 copies·μL−1、2.7×103 copies·μL−1分别进行3次组内、组间重复性试验,以判定该方法的重复性。
1.10 临床样品检测
对收集自福建地区猪场疑似PoRV的151份病料采用建立的方法进行检测和分析,并与RT-PCR方法进行对比,比较二者检测符合率。
2. 结果与分析
2.1 PoRV A质粒的鉴定
PoRV A质粒通过测序验证连接成功。通过NanoDrop2000测定PoRVA质粒浓度,根据质粒浓度计算公式[14],经计算可知质粒拷贝数为2.7×1010 copies·μL−1。PCR鉴定在598 bp处可见条带,与预期相符(图1)。测序结果显示,测序结果与目的序列一致。
2.2 反应体系优化
经条件优化后的荧光定量PCR体系:RT-qPCR Probe Mix 10.0 μL,模板 5.0 μL,RT Enzyme Mix 0.5 μL,引物PoRV- F3、PoRV- R3(10 μmol·L−1)各0.9 μL,探针(10 μmol·L−1)0.4 μL,RNase-free ddH2O 2.3 μL,总体系为 20.0 μL;程序:55 ℃ 15 min,95 ℃ 30 s;95 ℃ 15 s,60 ℃ 1 min(荧光采集),40个循环。
2.3 标准曲线的绘制
绘制的标准曲线如图2所示。PoRV A标准曲线线性方程为 y=−3.484x+40.086,相关系数R2达到
0.9991 ,扩增效率E为93.7%。结果表明,标准品模板的拷贝数与各自Ct值呈现良好的线性关系。2.4 特异性试验
利用优化后的TaqMan RT-qPCR反应体系和条件进行特异性检测。结果如图3所示,除PoRV A产生扩增荧光信号外,其他病毒和阴性对照均未检测到扩增信号,表明本试验建立的TaqMan荧光定量检测方法特异性强,不受其他常见猪源腹泻病毒影响。
2.5 敏感性试验
以10倍梯度稀释(2.7×101~2.7×108 copies·μL−1)的阳性质粒为模板,分别以建立的RT-qPCR方法和常规RT-PCR方法进行检测比较,其结果如图4和图5所示。 RT-qPCR检测方法的最低检测浓度为 2.7×101 copies·μL−1(以2.7×108~2.7×101 copies·μL−1为模板的RT-qPCR检测到的Ct值分别为9.22、11.83、15.20、18.60、21.26、24.79、27.62、31.92);常规RT-PCR方法的最低检测浓度为2.7×103 copies·μL−1。RT-qPCR检测灵敏度是RT-PCR灵敏度的100倍。
2.6 重复性试验
以 2.7×105 copies·μL−1、2.7×104 copies·μL−1、2.7×103 copies·μL−1的PoRV A质粒作为模板,应用建立的方法分别进行组内、组间重复试验。数据显示(表2),3个浓度均能有效扩增,该方法的组内变异系数为0.401%~0.932%;组间变异系数为0.359%~1.026%,说明该方法重复性好。
表 2 PoRV TaqMan RT-qPCR重复性试验结果Table 2. Results of PoRV TaqMan RT-qPCR assay质粒浓度
Concentration/
(copies·μL−1)组内变异试验
Intra-assay variability组间变异试验
Inter-assay variability平均数
¯X±SD变异系数
CV/%平均数
¯X±SD变异系数
CV/%2.7×105 21.553±0.201 0.932 22.507±0.231 1.026 2.7×104 24.413±0.098 0.401 24.853±0.127 0.511 2.7×103 27.177±0.248 0.912 27.540±0.099 0.359 2.7 临床样品检测
将收集到的151份来自福建省的疑似PoRV的临床样品分别使用TaqMan RT-qPCR与RT-PCR方法进行检测,结果如表3所示,TaqMan RT-qPCR的检出率为42.38%(64/151),可疑病料(Ct值35~40)有9份;RT-PCR的检出率为33.11%(50/151);荧光定量RT-PCR与RT-PCR结果符合率为78.16%(50/64)。
表 3 临床样品检测结果比较Table 3. Comparison of testing results on clinical specimens指标
Index阳性
Positive/份可疑
Suspicious/份阴性
Negative/份总计
Total/份RT-PCR 50 0 101 151 荧光定量 RT-PCR 64 9 78 151 3. 讨论与结论
猪腹泻类病毒对猪群危害大,临床监测重心一般为PEDV、PoRV、TGEV等病毒。PoRV A、PoRV B、PoRV C均可感染猪群,其中以PoRV A感染为主。感染PoRV的猪只通常会表现出腹泻、呕吐和脱水等典型症状。此外,轮状病毒感染的临床症状轻重取决于发病的日龄、免疫状态和环境条件,根据猪只的生长阶段不同,腹泻的具体表现也会有所差异。未满20日龄且缺乏母源抗体保护的仔猪,在感染PoRV后,会出现严重的水样腹泻,这导致仔猪严重脱水的同时死亡率增高。大于1月龄的仔猪在PoRV感染后虽然症状较轻,但也会出现腹泻呕吐的症状,对仔猪后续的生长发育造成影响[12]。妊娠期的母猪感染该病毒可能会导致流产和产出死胎。
凭借其高灵敏度和强特异性,实时荧光定量PCR技术在当前的检测条件下,能够实现对病原体快速且准确的检测。探针法与染料法相比,探针与扩增的目的序列的特异性结合产生荧光信号,不受引物二聚体的影响,也不受非特异性扩增的影响。因此,TaqMan荧光定量PCR技术已成为目前诊断多种病原体的关键方法。TaqMan荧光定量PCR与RT-PCR相比更加省时、快捷,并且实现病毒核酸定量[15]。采用一步法扩增,操作简单,同时避免了逆转录过程中需要频繁打开PCR管导致的核酸污染情况。
本研究以PoRV的VP6基因序列为基础,设计特异性引物与探针,建立了猪A群轮状病毒 TaqMan实时荧光定量PCR检测方法。目前VP6基因已经成为国内外研究轮状病毒的主要分子靶点,主要是由于VP6基因与其他基因片段相比变异程度小,序列相对保守[10]。特异性检测结果显示,该方法只特异性扩增PoRV A的核酸,对其他猪源腹泻类病毒均无扩增,表明该方法特异性强。敏感性试验结果显示,最低可以检测到浓度为27.0 copies·μL−1,敏感度是常规逆转录聚合酶链反应(RT-PCR)检测方法的100倍;与陈小飞等[16]建立的实时荧光定量PCR方法相比灵敏度更高,对于PoRV的早期诊断更具优势。重复性试验的结果表明,无论是组内试验还是组间试验,其变异系数均低于1.10%,这证实了该方法具有良好的稳定性和重复性。用该方法对收集自福建地区猪场疑似PoRV的151份病料进行检测结果显示,荧光定量RT-PCR方法的检出率为42.38%(64/151),逆转录聚合酶链反应(RT-PCR)方法的检出率为33.11%(50/151),说明该方法与常规逆转录聚合酶链反应(RT-PCR)的检测相比更加灵敏,同样说明PoRV是福建省猪场病毒性腹泻的病原之一,这值得我们更加重视PoRV的防控。
近些年来PoRV检出率逐渐增高,在我国湖北、河南、江苏、江西、浙江等地区均有报道,其检出率最高可达66.7%[4,17~20],国内PoRV流行病学调查报道2012年全国13个省份PoRV的感染率为3.20%[21];2021年左右,在何晓明等[22]的研究中,对我国6个省市的调查发现PoRV的感染率为27.89%;2023年关于PoRV的研究中发现江苏的PoRV阳性检出率达57.70%,江西地区的阳性检出率达55.71%[23],贵州的阳性检出率为38.79%[24]。本研究对福建地区151份病料的检测结果显示,PoRV的检出率为42.38%(64/151),与其他地区报道的数据相比,虽然不同地区的检出率存在差异,但检出率整体处于较高水平。
-
表 1 样本编码规则
Table 1 Sample codes
毛竹类型P. edulis type 样本编码 Sample encoding 竹鞭Rhizome 鞭根Rhizome root 根际土壤Rhizosphere soil 林间土壤Forest soil 大年毛竹IOn-year P. edulis I ON.1.A5 ON.1.B5 ON.1.C5 ON.CK.C5 大年毛竹IIOn-year P. edulis II ON.2.A5 ON.2.B5 ON.2.C5 大年毛竹IVOn-year P. edulis IV ON.4.A5 ON.4.B5 ON.4.C5 小年毛竹IOff-year P. edulis I OF.1.A5 OF.1.B5 OF.1.C5 OF.CK.C5 小年毛竹IIOff-year P. edulis II OF.2.A5 OF.2.B5 OF.2.C5 小年毛竹IVOff-year P. edulis IV OF.4.A5 OF.4.B5 OF.4.C5 表 2 各组样本的Alpha 多样性指数
Table 2 Alpha diversity index of samples
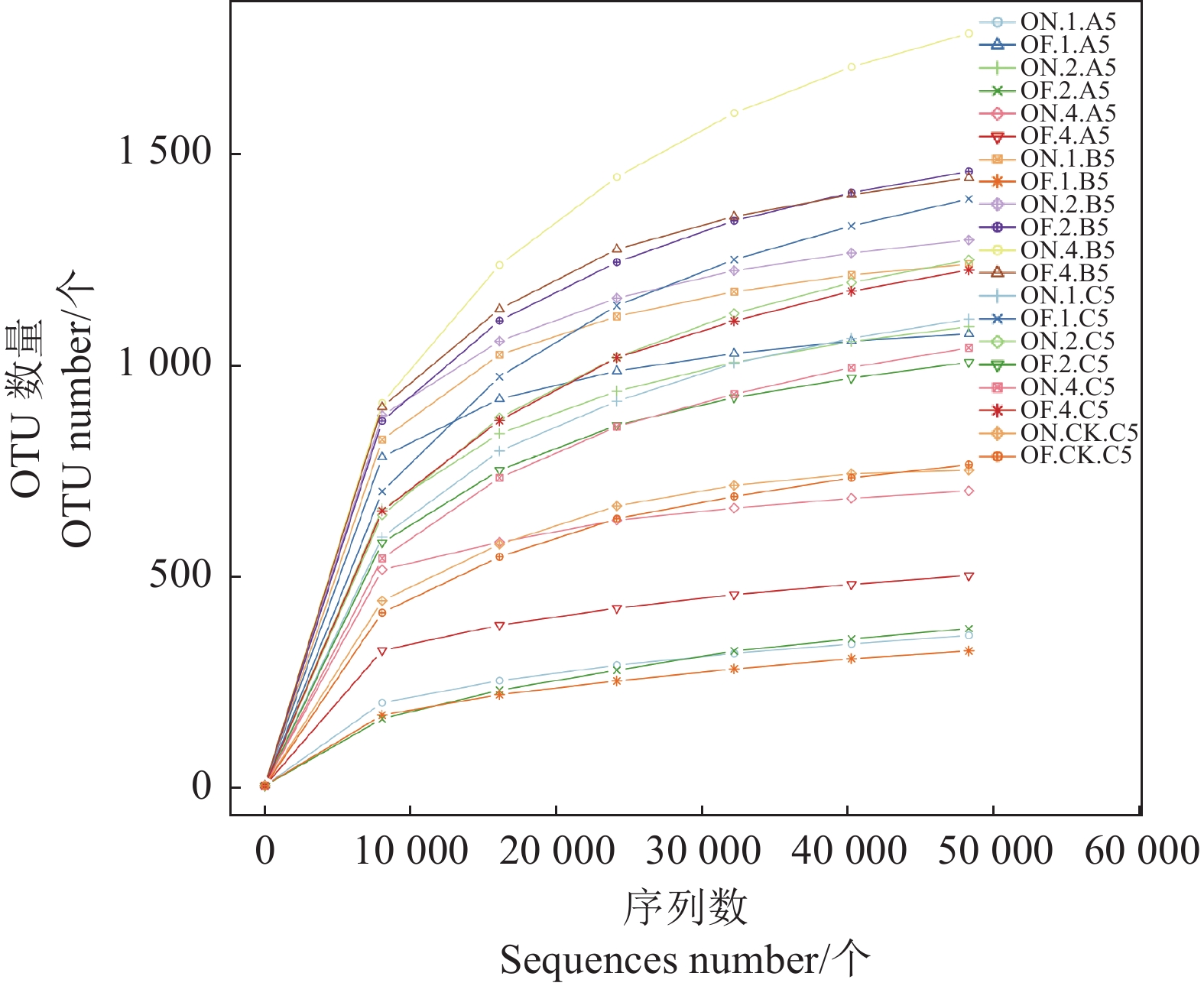
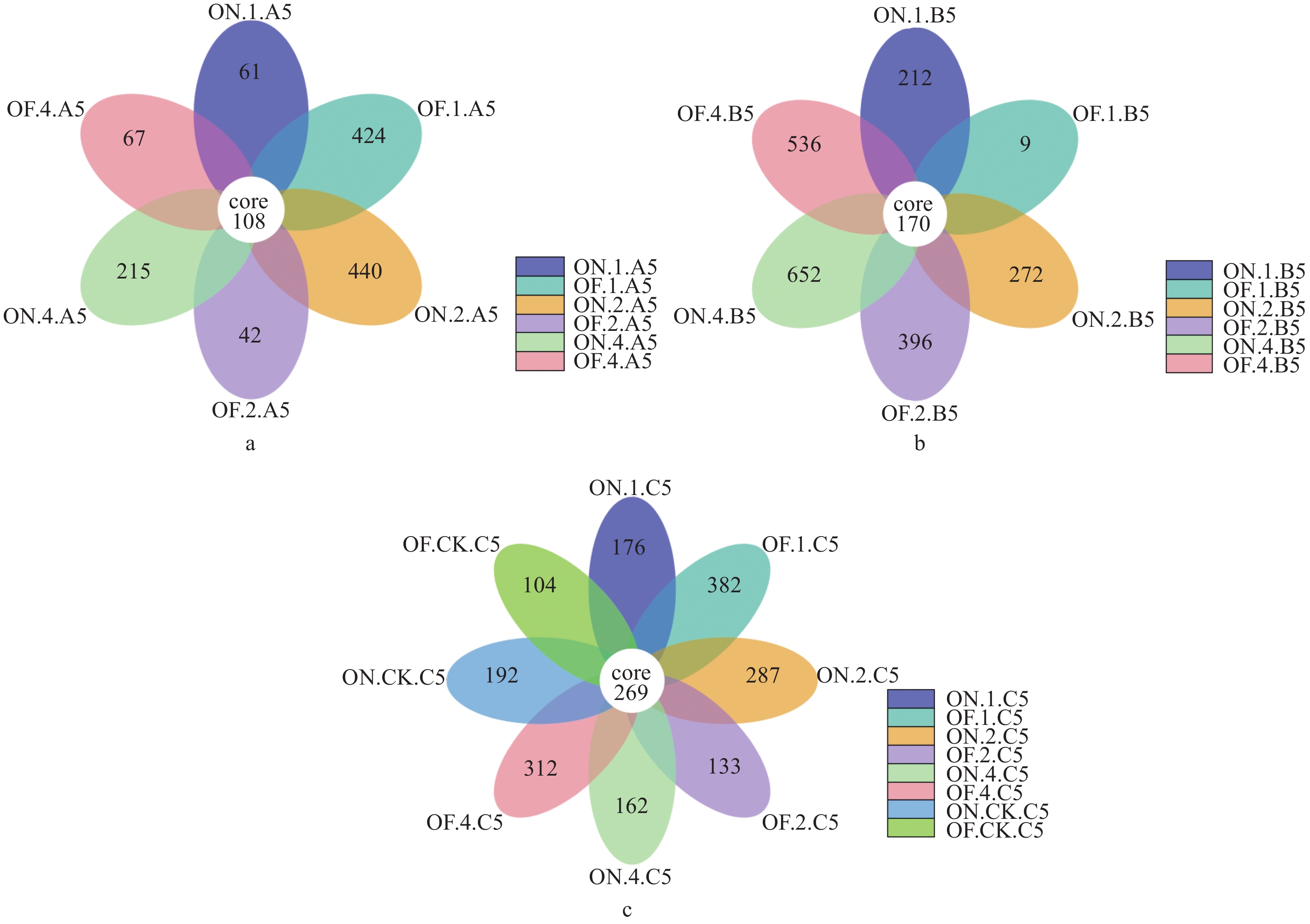
样本Sample 观测物种数/个Observed Species/piece 丰度指数ACE 香农指数Shannon 谱系多样性指数PD whole Tree 文库覆盖率Coverage/ % ON.1.A5 364 487.039 5.814 72.574 99.8 OF.1.A5 1078 1121.452 7.854 395.731 99.8 ON.2.A5 1095 1222.288 7.202 176.932 99.6 OF.2.A5 381 533.381 5.436 76.422 99.7 ON.4.A5 707 792.52 7.308 112.857 99.8 OF.4.A5 507 606.924 6.04 66.159 99.8 ON.1.B5 1243 1312.372 7.763 99.038 99.7 OF.1.B5 327 443.358 5.564 38.95 99.8 ON.2.B5 1300 1399.941 7.863 113.514 99.7 OF.2.B5 1462 1635.752 7.565 145.073 99.5 ON.4.B5 1790 2110.068 7.489 158.795 99.1 OF.4.B5 1447 1561.34 7.769 334.921 99.6 ON.1.C5 1113 1277.2 5.884 94.135 99.5 OF.1.C5 1397 1626.968 5.974 155.08 99.3 ON.2.C5 1253 1460.243 5.924 194.704 99.4 OF.2.C5 1011 1129.53 5.889 115.281 99.6 ON.4.C5 1045 1227.566 5.473 95.286 99.5 OF.4.C5 1229 1409.537 5.54 109.603 99.5 ON.CK.C5 756 756 5.776 95.302 100 OF.CK.C5 769 877.076 5.082 67.327 99.7 -
[1] PENG Z H, LU Y, LI L B, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) [J]. Nature Genetics, 2013, 45(4): 456−461. DOI: 10.1038/ng.2569
[2] LONGWEI, LI N, LU D S, et al. Mapping Moso bamboo forest and its on-year and off-year distribution in a subtropical region using time-series Sentinel-2 and Landsat 8 data [J]. Remote Sensing of Environment, 2019, 231: 111265. DOI: 10.1016/j.rse.2019.111265
[3] ZHOU Y F, ZHOU G M, DU H Q, et al. Biotic and abiotic influences on monthly variation in carbon fluxes in on-year and off-year Moso bamboo forest [J]. Trees, 2019, 33(1): 153−169. DOI: 10.1007/s00468-018-1765-1
[4] SHELAKE R M, PRAMANIK D, KIM J Y. Exploration of plant-microbe interactions for sustainable agriculture in CRISPR era [J]. Microorganisms, 2019, 7(8): 269. DOI: 10.3390/microorganisms7080269
[5] MÜLLER D B, VOGEL C, BAI Y, et al. The plant microbiota: Systems-level insights and perspectives [J]. Annual Review of Genetics, 2016, 50: 211−234. DOI: 10.1146/annurev-genet-120215-034952
[6] MA B, WANG H Z, DSOUZA M, et al. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in Eastern China [J]. The ISME Journal, 2016, 10(8): 1891−1901. DOI: 10.1038/ismej.2015.261
[7] KLOEPPER J W, LEONG J, TEINTZE M, et al . Enhanced plant growth by siderophores produced by plant growth -promothing rhizobacteria [J]. Nature, 1980, 286 (5776): 885−886 . DOI: 10.1038/286885a0
[8] BULGARELLI D, SCHLAEPPI K, SPAEPEN S, et al . Sturcture and functions of the bacterial microbiota of plant [J]. Annual Review of Plant Biology, 2013, 64 : 807−838. DOI: 10.1146/annurev-arplant-050312-120106
[9] 吴良如,萧江华. 大小年毛竹林中内源激素节律变化特征的研究 [J]. 竹子研究汇刊,1998, 1998, 17(1):24−30. LIANGU W, JIANGHUA XIAO. Study on Dynamic Characteristics of Eudogenous Phytohormone in On-and-Off Year Bamboo (Phyllost achys Heterocycles Var. Pubescens) Grove [J]. Journal of Bamboo Research, 1998, 17(1): 24−30.(in Chinese)
[10] EDGAR R C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads [J]. Nature Methods, 2013, 10(10): 996−998. DOI: 10.1038/nmeth.2604
[11] LIN X C, CHOW T Y, CHEN H H, et al. Understanding bamboo flowering based on large-scale analysis of expressed sequence tags [J]. Genetics and Molecular Research:GMR, 2010, 9(2): 1085−1093. DOI: 10.4238/vol9-2gmr804
[12] SCHLOSS P D, WESTCOTT S L, RYABIN T, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities [J]. Applied and Environmental Microbiology, 2009, 75(23): 7537−7541. DOI: 10.1128/AEM.01541-09
[13] ISAGI Y, SHIMADA K, KUSHIMA H, et al. Clonal structure and flowering traits of a bamboo[Phyllostachys pubescens (Mazel) Ohwi]stand grown from a simultaneous flowering as revealed by AFLP analysis [J]. Molecular Ecology, 2004, 13(7): 2017−2021. DOI: 10.1111/j.1365-294X.2004.02197.x
[14] BAIS H P, WEIR T L, PERRY L G, et al. The role of root exudates in rhizosphere interactions with plants and other organisms [J]. Annual Review of Plant Biology, 2006, 57: 233−266. DOI: 10.1146/annurev.arplant.57.032905.105159
[15] REINHOLD-HUREK B, BÜNGER W, BURBANO C S, et al. Roots shaping their microbiome: Global hotspots for microbial activity [J]. Annual Review of Phytopathology, 2015, 53: 403−424. DOI: 10.1146/annurev-phyto-082712-102342
[16] COLEMAN-DERR D, DESGARENNES D, FONSECA-GARCIA C, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species [J]. The New Phytologist, 2016, 209(2): 798−811. DOI: 10.1111/nph.13697
[17] XUAN D T, GUONG V T, ROSLING A, et al. Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa [J]. Biology and Fertility of Soils, 2012, 48(2): 217−225. DOI: 10.1007/s00374-011-0618-5
[18] XIANGZHEN, LI,. Dynamics of the bacterial community structure in the rhizosphere of a maize cultivar [J]. Soil Biology and Biochemistry, 2014, 68: 392−401. DOI: 10.1016/j.soilbio.2013.10.017
[19] SHI Y H, PAN Y S, XIANG L, et al. Assembly of rhizosphere microbial communities in Artemisia annua: Recruitment of plant growth-promoting microorganisms and inter-Kingdom interactions between bacteria and fungi [J]. Plant and Soil, 2022, 470(1): 127−139.
[20] BERENDSEN R L, PIETERSE C M J, BAKKER P A H M. The rhizosphere microbiome and plant health [J]. Trends in Plant Science, 2012, 17(8): 478−486. DOI: 10.1016/j.tplants.2012.04.001
[21] HENNING S M, YANG J P, SHAO P, et al. Health benefit of vegetable/fruit juice-based diet: Role of microbiome [J]. Scientific Reports, 2017, 7: 2167. DOI: 10.1038/s41598-017-02200-6
[22] PENG G X, ZHANG W, LUO H F, et al. Enterobacter oryzae sp. nov. , a nitrogen-fixing bacterium isolated from the wild rice species Oryza latifolia[J]. International Journal of Systematic and Evolutionary Microbiology, 2009, 59(Pt 7): 1650-1655.
[23] BOTHE H. Biology of the Nitrogen Cycle[M]. Amsterdam: Elsevier Science Ltd, 2007: 147-163.
[24] OROZCO-MOSQUEDA M D C, ROCHA-GRANADOS M D C, GLICK B R, et al. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms [J]. Microbiological Research, 2018, 208: 25−31. DOI: 10.1016/j.micres.2018.01.005
[25] STÉPHANE, COMPANT C, SESSITSCH A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization [J]. Soil Biology and Biochemistry, 2010, 42(5): 669−678. DOI: 10.1016/j.soilbio.2009.11.024
[26] 张爱梅, 殷一然, 孙坤. 沙棘属植物弗兰克氏菌研究进展 [J]. 微生物学通报, 2020, 47(11):3933−3944. DOI: 10.13344/j.microbiol.china.200427 ZHANG A M, YIN Y R, SUN K. Research progress in Frankia spp. associated with Hippophae L [J]. Microbiology China, 2020, 47(11): 3933−3944.(in Chinese) DOI: 10.13344/j.microbiol.china.200427
[27] DIAGNE N, ARUMUGAM K, NGOM M, et al. Use of Frankia and actinorhizal plants for degraded lands reclamation [J]. BioMed Research International, 2013, 2013: 948258.
[28] 黄瑞林, 张娜, 孙波, 等. 典型农田根际土壤伯克霍尔德氏菌群落结构及其多样性 [J]. 土壤学报, 2020, 57(4):975−985. DOI: 10.11766/trxb201901040008 HUANG R L, ZHANG N, SUN B, et al. Community structure of burkholderiales and its diversity in typical maize rhizosphere soil [J]. Acta Pedologica Sinica, 2020, 57(4): 975−985.(in Chinese) DOI: 10.11766/trxb201901040008
[29] SIJAM K, DIKIN A. Biochemical and physiological characterization of Burkholderia cepacia as biological control agent [J]. International Journal of Agriculture & Biology, 2005, 7(3): 385−388.
[30] HARDOIM P R, VAN OVERBEEK L S, ELSAS J D. Properties of bacterial endophytes and their proposed role in plant growth [J]. Trends in Microbiology, 2008, 16(10): 463−471. DOI: 10.1016/j.tim.2008.07.008
[31] LEBEIS S L. The potential for give and take in plant-microbiome relationships [J]. Frontiers in Plant Science, 2014, 5: 287.




 下载:
下载: