Screening and analysis on reference genes of Ganoderma pseudoferreum
-
摘要:
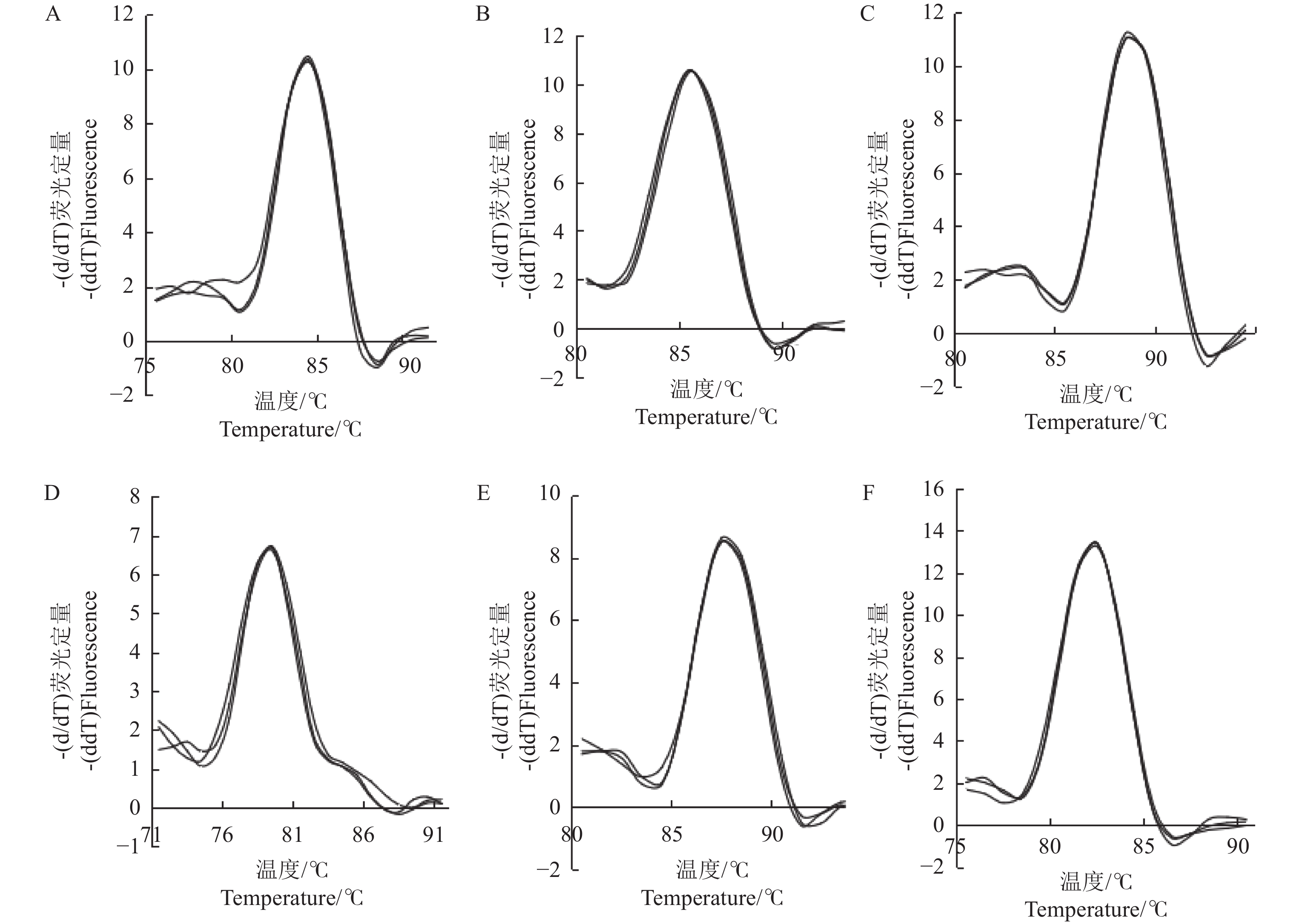
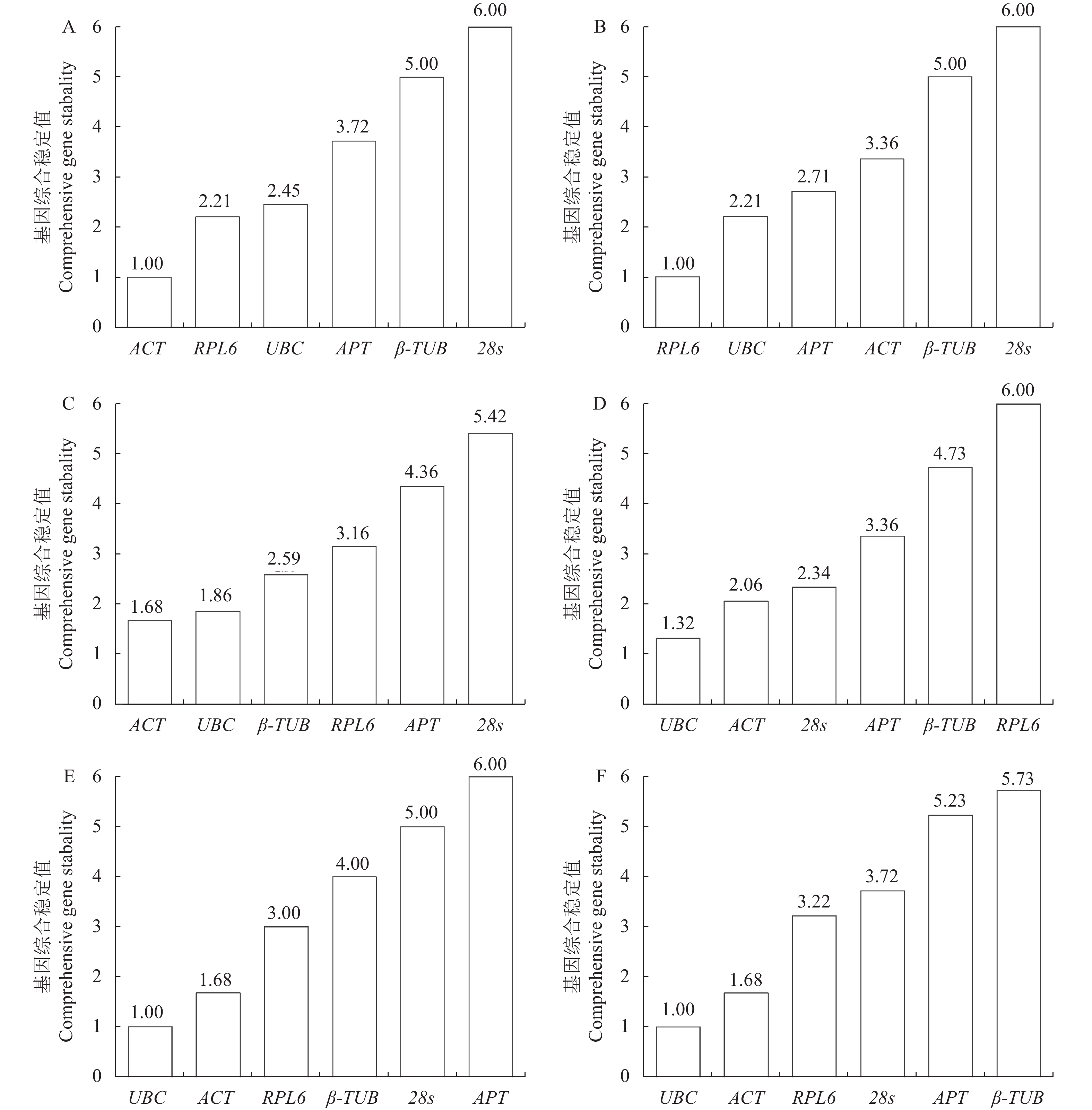
目的 分析评价不同胁迫处理的橡胶灵芝菌候选内参基因的稳定性,以期为探索橡胶灵芝菌基因的功能及其侵染橡胶树的分子机制等提供参考。 方法 以5个非生物因子(温度、盐、氧化、pH、干旱)和1个生物因子(生防细菌)胁迫下的橡胶灵芝菌作为材料,提取RNA并反转录为cDNA,采用实时荧光定量PCR技术,扩增6个候选内参基因(UBC、ACT、RPL6、β-TUB、APT、28s),利用分析软件geNorm、NormFinder、BestKeeper和RefFinder根据基因表达的稳定性对基因进行排序,选择最适合不同胁迫的内参基因组合。 结果 所有样品RNA均为清晰的两条带,且6个候选内参基因的熔解曲线为明显单一峰。结合geNorm、NormFinder、Bestkeeper和RefFinder对其进行表达稳定性分析发现,温度胁迫下基因表达稳定性为UBC>ACT>RPL6>β-TUB>28s>APT,盐胁迫下基因表达稳定性为ACT>RPL6>UBC>APT>β-TUB>28s;氧化胁迫下基因表达稳定性为UBC>ACT>28s>APT>β-TUB>RPL6;pH胁迫下基因表达稳定性为RPL6>UBC>APT>ACT>β-TUB>28s,干旱胁迫下基因表达稳定性为ACT>UBC>β-TUB>RPL6>APT>28s,生物胁迫下基因表达稳定性为UBC>ACT>RPL6>28s>APT>β-TUB。 结论 结合所有候选基因稳定性和胁迫条件,推荐基因ACT和UBC作为在干旱、氧化、温度和生物胁迫下的内参基因,基因ACT和RPL6为盐胁迫下的内参基因,UBC和RPL6为pH胁迫下的内参基因。本研究结果为不同胁迫下橡胶灵芝菌相关基因的表达研究提供了合适的内参基因。 Abstract: :Objective The stability of candidate reference genes of Ganoderma pseudoferreum under different stress treatments were evaluated to provide reference for exploring the gene function and molecular mechanism of G.pseudoferreum infestation of rubber tree roots. Methods The G. pseudoferreum mycelia under five abiotic stresses ( temperature, salt, oxidation, pH, drought ) and one biotic stress (biocontrol bacteria) were collected to isolate RNA and reverse transcribed into cDNA. Real-time fluorescence quantitative PCR technology was used to amplify the six candidate reference genes (UBC, ACT, RPL6, β-TUB, APT, 28s), and the softwares of geNorm, NormFinder, BestKeeper and RefFinder were used to evaluate genes stability to find the most suitable reference genes for different stresses. Results The RNA of all samples showed two clear bands, and the melting curves of the six candidate reference genes showed obvious single peak. Combined analysis of geNorm, NormFinder, Bestkeeper and RefFinder, found that the stability of gene expression under temperature stress was UBC > ACT > RPL6 > β-TUB > 28s > APT, and the stability of gene expression under salt stress was ACT > RPL6 > UBC > APT > β-TUB > 28s; the stability of gene expression under oxidative stress was UBC > ACT > 28s > APT > β-TUB > RPL6; the stability of gene expression under pH stress was RPL6 > UBC > APT > ACT > β-TUB > 28s, the stability of gene expression under drought stress was ACT > UBC > β-TUB > RPL6 > APT > 28s, and the stability of gene expression under biotic stress was UBC > ACT > RPL6 > 28s > APT > β-TUB. Conclusion Combining all genes and experimental conditions, we recommend genes ACT and UBC as the best reference genes for normalizing gene expression under drought, oxidation, temperature and biotic stress experimental conditions. Genes ACT and RPL6 are the most suitable reference genes under salt stress, and UBC and RPL6 are the most suitable reference genes under pH stress. -
Key words:
- Rubber tree /
- Red root rot disease /
- Ganoderma pseudoferreum /
- Reference genes /
- RT-qPCR
-
表 1 RT-qPCR引物
Table 1. RT-qPCR primers
基因
GeneGene Bank登录号
Gene Bank accessing number引物序列
Primer sequence (5'-3')扩增长度/bp
Amplification length /bpUBC KAF9027008 TCTGGCGGCGTCTTCTTCCT 106 TGGCATTGATGTTCGGGTGG ACT KAF9050787 CATCGAGCACGGTATTGTCA 167 TCTCGAACATGATTTGGGTC β-TUB KAF9039628 CAAATGCAGAACGTCCAGAAC 159 GTGAACTCCATCTCGTCCATAC RPL6 KAF9033237 CTGTACCTCGTCGGTGTCGG 137 GTTGGCGTCTCCACCTTTGC APT GL18178 GAGTACGGTGTGGATGTCTTC 130 CGAGCTTGGCTACGAGTTC 28s 无登录号 GCATATCAATAAGCGGAGGA 130 GCACTTCTCCAGACTACAAC 表 2 NormFinder稳定性分析
Table 2. Stability Analysis by NormFinder
排名
Rank盐胁迫
Salt stress (GroupSD)pH胁迫
pH stress (GroupSD)干旱胁迫
Drought stress (GroupSD)氧化胁迫
Oxidative stress (GroupSD)温度胁迫
Heat stress (GroupSD)生物胁迫
Biotic stress (GroupSD)1 RPL6 RPL6 UBC UBC UBC UBC GroupSD 0.02 0.05 0.05 0.02 0.04 0.04 2 APT β-TUB ACT APT ACT ACT GroupSD 0.02 0.05 0.05 0.02 0.04 0.06 3 UBC APT β-TUB ACT RPL6 RPL6 GroupSD 0.11 0.05 0.07 0.02 0.12 0.07 4 β-TUB UBC RPL6 RPL6 β-TUB APT GroupSD 0.11 0.07 0.07 0.1 0.19 0.08 5 ACT ACT APT β-TUB 28s 28s GroupSD 0.11 0.07 0.15 0.11 0.46 0.11 6 28s 28s 28s 28s APT β-TUB GroupSD 0.53 0.24 0.24 0.15 0.56 0.12 表 3 Bestkeeper稳定性分析
Table 3. Stability Analysis by Bestkeeper
排名
Rank盐胁迫
Salt stress (CV±SD)pH胁迫
pH stress (CV±SD)干旱胁迫
Drought stress (CV±SD)氧化胁迫
Oxidative stress (CV±SD)温度胁迫
Heat stress (CV±SD)生物胁迫
Biotic stress (CV±SD)1 ACT RPL6 UBC UBC UBC UBC CV±SD 2.17±0.37 0.89±0.25 1.53±0.23 2.90±0.37 0.44±0.06 0.53±0.07 2 RPL6 APT RPL6 ACT RPL6 ACT CV±SD 2.30±0.65 1.11±0.26 1.84±0.49 3.29±1.46 1.32±0.44 1.11±0.20 3 UBC ACT ACT APT ACT RPL6 CV±SD 3.02±0.50 1.71±0.30 2.50±0.42 3.59±0.82 3.72±0.48 1.50±0.43 4 β-TUB UBC β-TUB β-TUB β-TUB β-TUB CV±SD 3.07±0.74 1.74±0.29 4.23±1.07 3.86±1.21 4.35±1.03 4.46±1.45 5 APT β-TUB APT RPL6 28s APT CV±SD 3.68±0.87 2.73±0.67 5.19±1.23 5.47±1.45 25.65±4.14 6.79±1.72 6 28s 28s 28s 28s APT 28s CV±SD 25.41±1.95 23.73±1.88 10.06±1.21 6.82±0.59 32.66±5.95 7.41±0.97 -
[1] NANDRIS D, NICOLE M, GEIGER J P. Root rot diseases of rubber trees [J]. Plant Disease, 1987, 71(4): 298−306. doi: 10.1094/PD-71-0298 [2] 张运强, 张辉强, 邓晓东. 橡胶树红根病病原菌的鉴定 [J]. 热带作物学报, 1997, 18(1):16−23.ZHANG Y Q, ZHANG H Q, DENG X D. Identification of pathogenic fungi of rubber red root disease [J]. Chinese Journal of Tropical Crops, 1997, 18(1): 16−23. (in Chinese) [3] 丁婧钰. 橡胶树与相思树病原灵芝种类鉴定及生物学特性研究[D]. 海口: 海南大学, 2018.DING J Y. Type identification of pathogenic Ganoderma and biological characteristics of the rubber tree and Acacia spp. [D]. Haikou: Hainan University, 2018. (in Chinese) [4] YANG Y T, ZHANG X, CHEN Y, et al. Selection of reference genes for normalization of microRNA expression by RT-qPCR in sugarcane buds under cold stress [J]. Frontiers in Plant Science, 2016, 7: 86. [5] ZHAO X, YANG H L, CHEN M J, et al. Reference gene selection for quantitative real-time PCR of mycelia from Lentinula edodes under high-temperature stress [J]. BioMed Research International, 2018, 2018: 1670328. [6] 张越. 黑木耳qRT-PCR内参基因的筛选及功能基因表达水平的研究[D]. 长春: 吉林农业大学, 2020.ZHANG Y. Studies of screening of reference genes and functional genes expression levels of Auricularia heimuer for qRT-PCR[D]. Changchun: Jilin Agricultural University, 2020. (in Chinese) [7] 刘英, 喻晓明, 蔡佺佑, 等. 巨大口蘑内参基因的筛选 [J]. 食用菌学报, 2017, 24(4):12−18.LIU Y, YU X M, CAI Q Y, et al. Reference gene selection for real-time quantitative PCR in Tricholoma giganteum [J]. Acta Edulis Fungi, 2017, 24(4): 12−18. (in Chinese) [8] 赵建霞, 沈颖越, 冯伟林, 等. 双孢蘑菇内参基因的筛选与矫正 [J]. 浙江农业学报, 2019, 31(8):1312−1320. doi: 10.3969/j.issn.1004-1524.2019.08.12ZHAO J X, SHEN Y Y, FENG W L, et al. Screening of internal reference gene of Agaricus bisporus [J]. Acta Agriculturae Zhejiangensis, 2019, 31(8): 1312−1320. (in Chinese) doi: 10.3969/j.issn.1004-1524.2019.08.12 [9] XU J, XU Z C, ZHU Y J, et al. Identification and evaluation of reference genes for qRT-PCR normalization in Ganoderma lucidum [J]. Current Microbiology, 2014, 68(1): 120−126. doi: 10.1007/s00284-013-0442-2 [10] 武晨剑. 金针菇不同发育阶段差异基因筛选及转录因子FfMYB和FfGAL基因的原核表达与纯化[D]. 太谷: 山西农业大学, 2021.WU C J. Screening of differentially expression genes related to different developmental stages of Flammulina filiformis and prokaryotic expression and purification of transcription factors FfMYB and FfGAL[D]. Taigu: Shanxi Agricultural University, 2021. (in Chinese) [11] ANDERSEN C L, JENSEN J L, ØRNTOFT T F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets [J]. Cancer Research, 2004, 64(15): 5245−5250. doi: 10.1158/0008-5472.CAN-04-0496 [12] VANDESOMPELE J, DE PRETER K, PATTYN F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes [J]. Genome Biology, 2002, 3(7): RESEARCH0034. doi: 10.1186/gb-2002-3-7-reports0034 [13] PFAFFL M W, TICHOPAD A, PRGOMET C, et al. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations [J]. Biotechnology Letters, 2004, 26(6): 509−515. doi: 10.1023/B:BILE.0000019559.84305.47 [14] DHEDA K, HUGGETT J F, CHANG J S, et al. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization [J]. Analytical Biochemistry, 2005, 344(1): 141−143. doi: 10.1016/j.ab.2005.05.022 [15] XIE F L, XIAO P, CHEN D L, et al. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs [J]. Plant Molecular Biology, 2012, 80(1): 75−84. doi: 10.1007/s11103-012-9885-2 [16] 孙茜茜, 何其光, 靳鹏飞, 等. 橡胶树红根病菌GPFTR1基因的克隆、亚细胞定位与表达分析 [J]. 基因组学与应用生物学, 2019, 38(8):3646−3653.SUN Q Q, HE Q G, JIN P F, et al. Cloning, subcellular localization and expression analysis of GPFTR1 gene from Ganoderma pseudoferreum [J]. Genomics and Applied Biology, 2019, 38(8): 3646−3653. (in Chinese) [17] 伏雪, 涂敏, 蔡海滨, 等. 橡胶树红根病病原菌木聚糖酶编码基因GpTR1774的克隆与表达分析 [J]. 热带作物学报, 2023, 44(12):2461−2468. doi: 10.3969/j.issn.1000-2561.2023.12.011FU X, TU M, CAI H B, et al. Cloning and expression analysis of xylanase GpTR1774 gene from Ganoderma pseudoferreum [J]. Chinese Journal of Tropical Crops, 2023, 44(12): 2461−2468. (in Chinese) doi: 10.3969/j.issn.1000-2561.2023.12.011 [18] 林仕恺, 涂敏, 鲁红学, 等. 巴西橡胶树红根病病菌液体培养条件优化研究 [J]. 热带农业科学, 2014, 34(10):71−74. doi: 10.3969/j.issn.1009-2196.2014.10.016LIN S K, TU M, LU H X, et al. Optimization of Ganoderma pseudoferreum liquid culture condition [J]. Chinese Journal of Tropical Agriculture, 2014, 34(10): 71−74. (in Chinese) doi: 10.3969/j.issn.1009-2196.2014.10.016 [19] CAO L P, ZHANG Q, MIAO R Y, et al. Reference gene selection for quantitative real-time PCR analysis of Hymenopellis radicata under abiotic stress [J]. Fungal Biology, 2024, 128(1): 1567−1577. doi: 10.1016/j.funbio.2023.11.004 [20] 吴建阳, 何冰, 杜玉洁, 等. 利用geNorm、NormFinder和BestKeeper软件进行内参基因稳定性分析的方法 [J]. 现代农业科技, 2017, (5):278−281. doi: 10.3969/j.issn.1007-5739.2017.05.174WU J Y, HE B, DU Y J, et al. Analysis method of systematically evaluating stability of reference genes using geNorm, NormFinder and BestKeeper [J]. Modern Agricultural Science and Technology, 2017(5): 278−281. (in Chinese) doi: 10.3969/j.issn.1007-5739.2017.05.174 [21] 张晓华, 孙达锋, 刘绍雄, 等. 食(药)用菌内参基因筛选研究进展 [J]. 中国食用菌, 2023, 42(5):1−6.ZHANG X H, SUN D F, LIU S X, et al. Research progress of internal reference gene screening for edible and medicinal fungi [J]. Edible Fungi of China, 2023, 42(5): 1−6. (in Chinese) [22] JAIN M, NIJHAWAN A, TYAGI A K, et al. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR [J]. Biochemical and Biophysical Research Communications, 2006, 345(2): 646−651. doi: 10.1016/j.bbrc.2006.04.140 [23] FANG P, LU R F, SUN F, et al. Assessment of reference gene stability in Rice stripe virus and Rice black streaked dwarf virus infection rice by quantitative Real-time PCR [J]. Virology Journal, 2015, 12: 175. doi: 10.1186/s12985-015-0405-2 [24] HERIDE C, URBÉ S, CLAGUE M J. Ubiquitin code assembly and disassembly [J]. Current Biology, 2014, 24(6): R215−R220. doi: 10.1016/j.cub.2014.02.002 [25] XIANG Q J, LI J, QIN P, et al. Identification and evaluation of reference genes for qRT-PCR studies in Lentinula edodes [J]. PLoS One, 2018, 13(1): e0190226. doi: 10.1371/journal.pone.0190226 [26] WEI Y M, LIU Y, LI L, et al. Identification of s9ap used as an endogenous reference gene in qualitative and real-time quantitative PCR detection of Pleurotus eryngii [J]. Molecular Biology Reports, 2023, 50(1): 621−629. doi: 10.1007/s11033-022-07562-3 [27] QIAN J, GAO Y N, WÁNG Y, et al. Selection and evaluation of appropriate reference genes for RT-qPCR normalization of Volvariella volvacea gene expression under different conditions [J]. BioMed Research International, 2018, 2018: 6125706. [28] ZHANG C, LI T, HOU C L, et al. Selection of reference genes from Shiraia bambusicola for RT-qPCR analysis under different culturing conditions [J]. AMB Express, 2017, 7(1): 14. doi: 10.1186/s13568-016-0314-9 -








 下载:
下载: