Full-length Transcriptome of She Herbal Medicine Plant Clematis florida
-
摘要:目的
重瓣铁线莲是畲药十二时辰基源植物,以根入药,含有丰富的活性成分。本研究旨在了解重瓣铁线莲基因注释信息、次级代谢产物代谢通路及基因功能,丰富其转录组信息,为进一步筛选和鉴定其药用成分代谢通路相关基因奠定基础。
方法利用SMRT测序技术,对重瓣铁线莲根进行全长转录组测序,并借助生物信息学工具进行功能注释、结构分析以及萜类合成途径基因的挖掘。
结果测序共获得高质量测序subreads数据量62.21 Gb,通过数据处理分析,获得
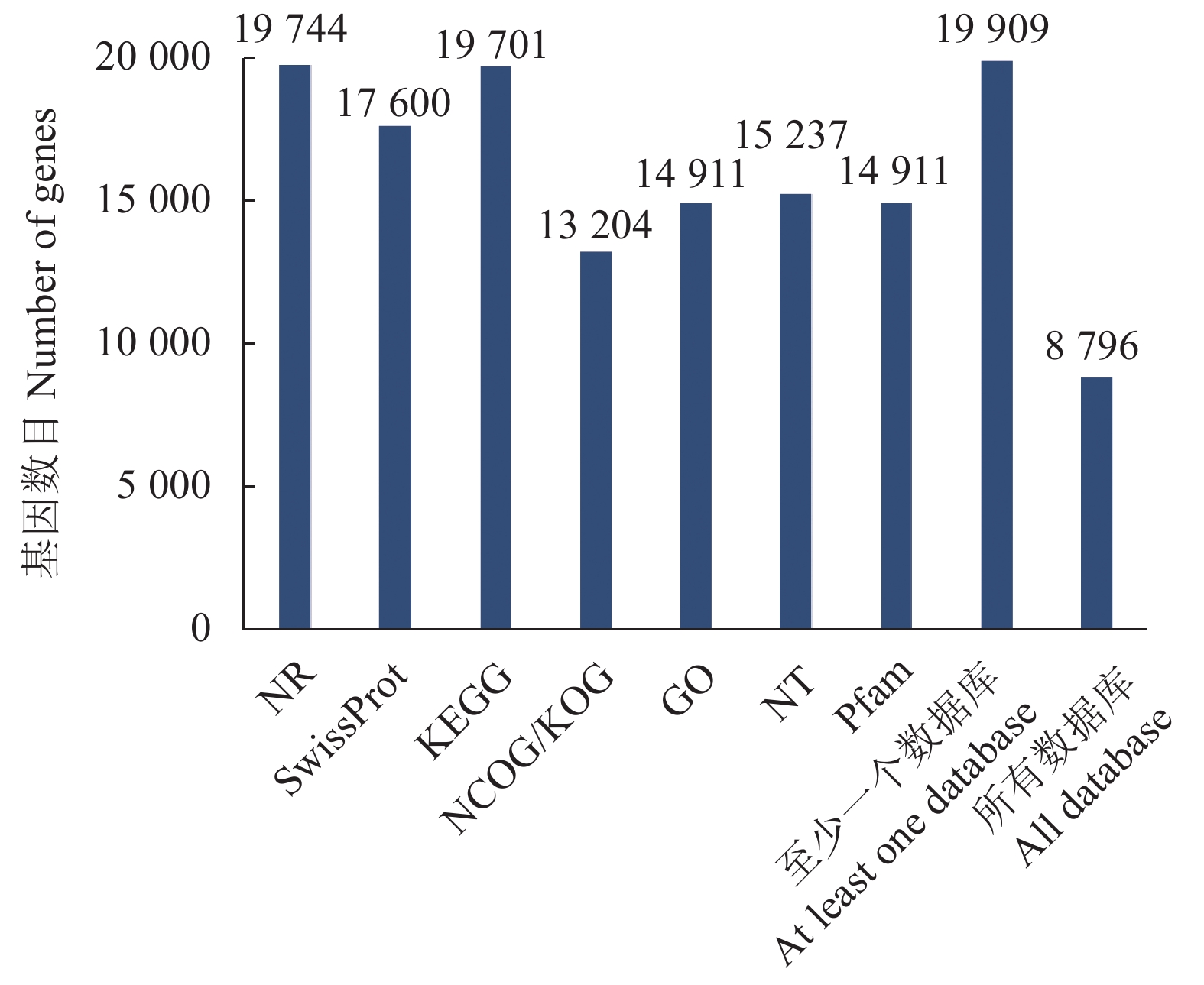
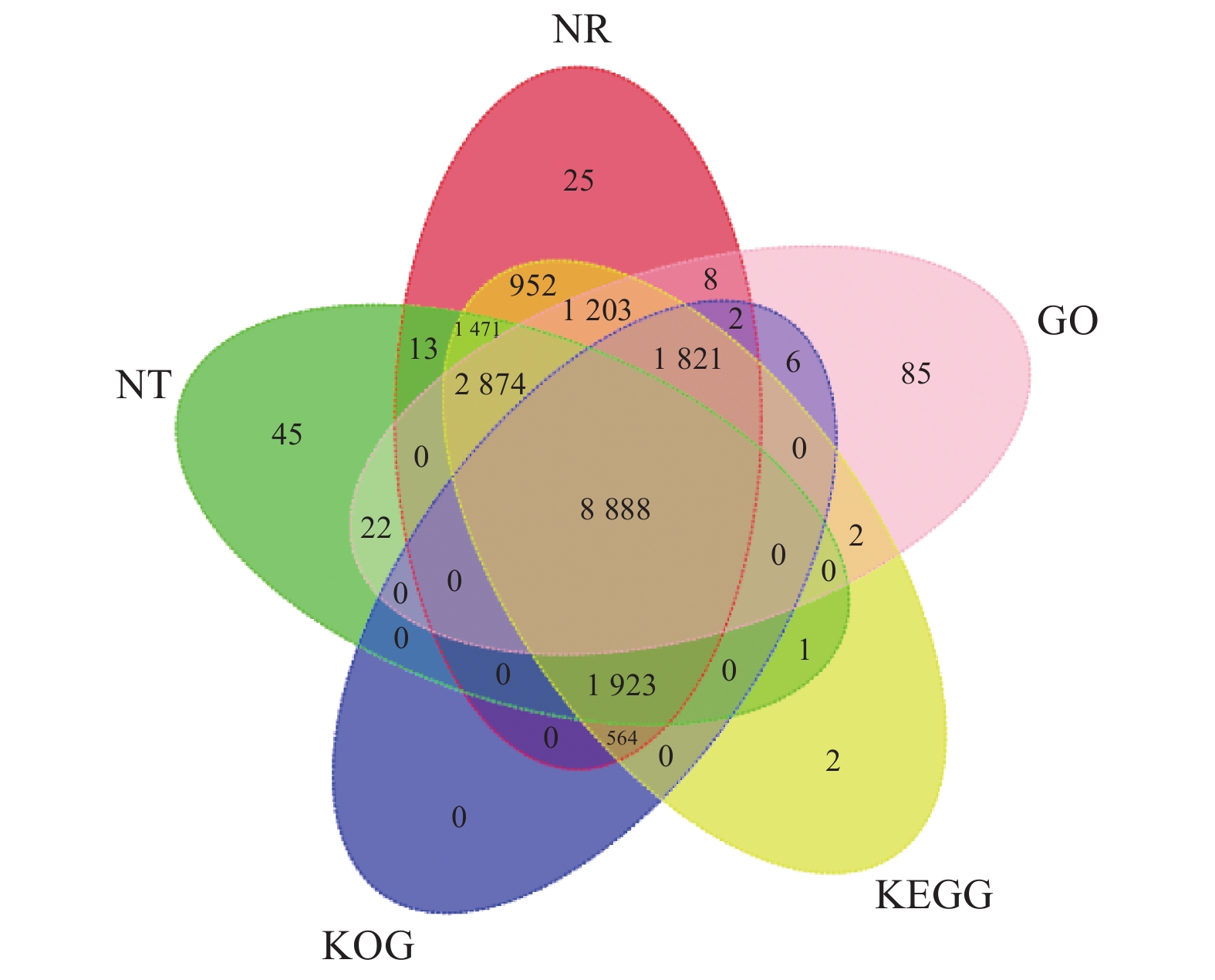
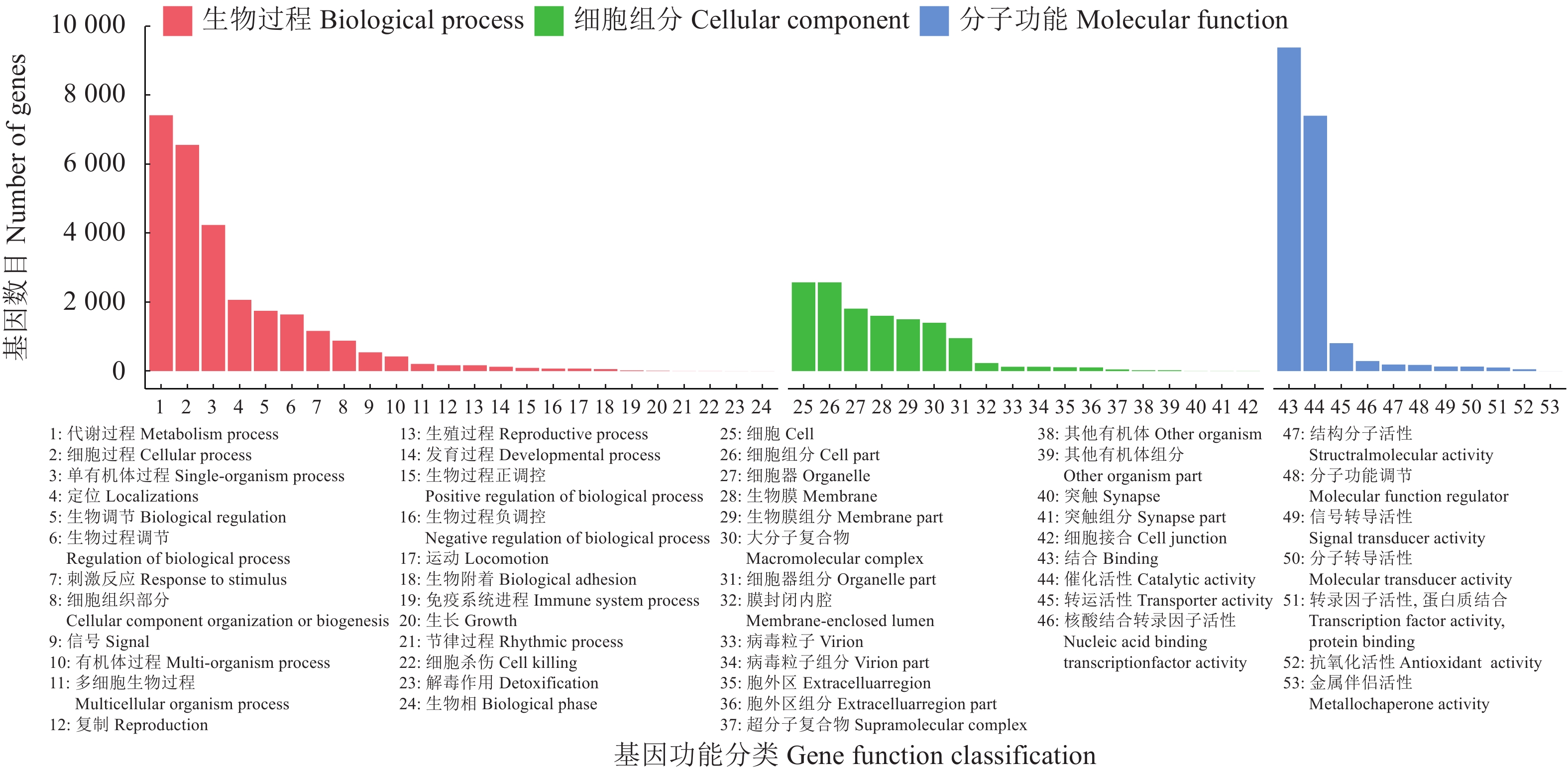
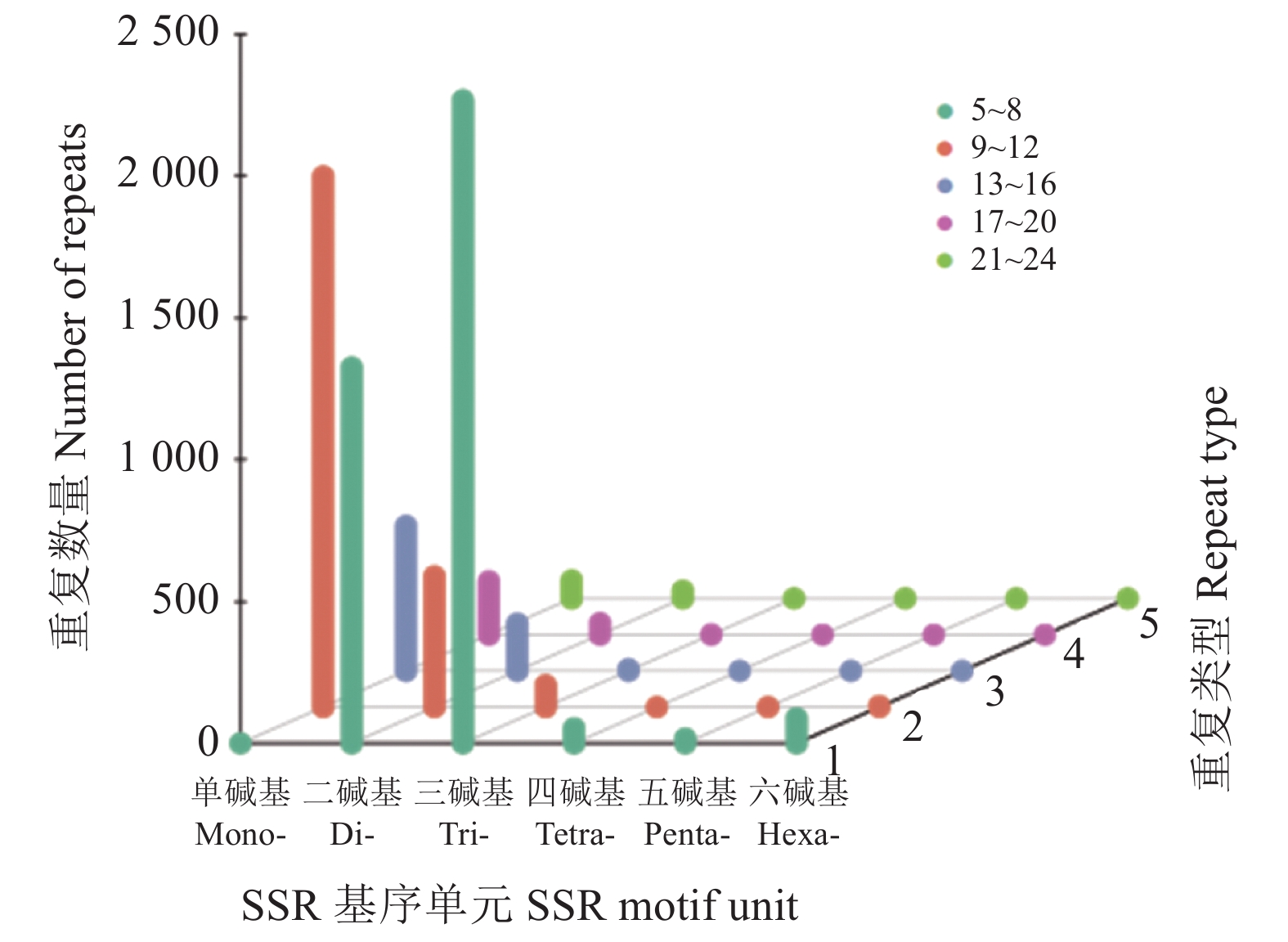
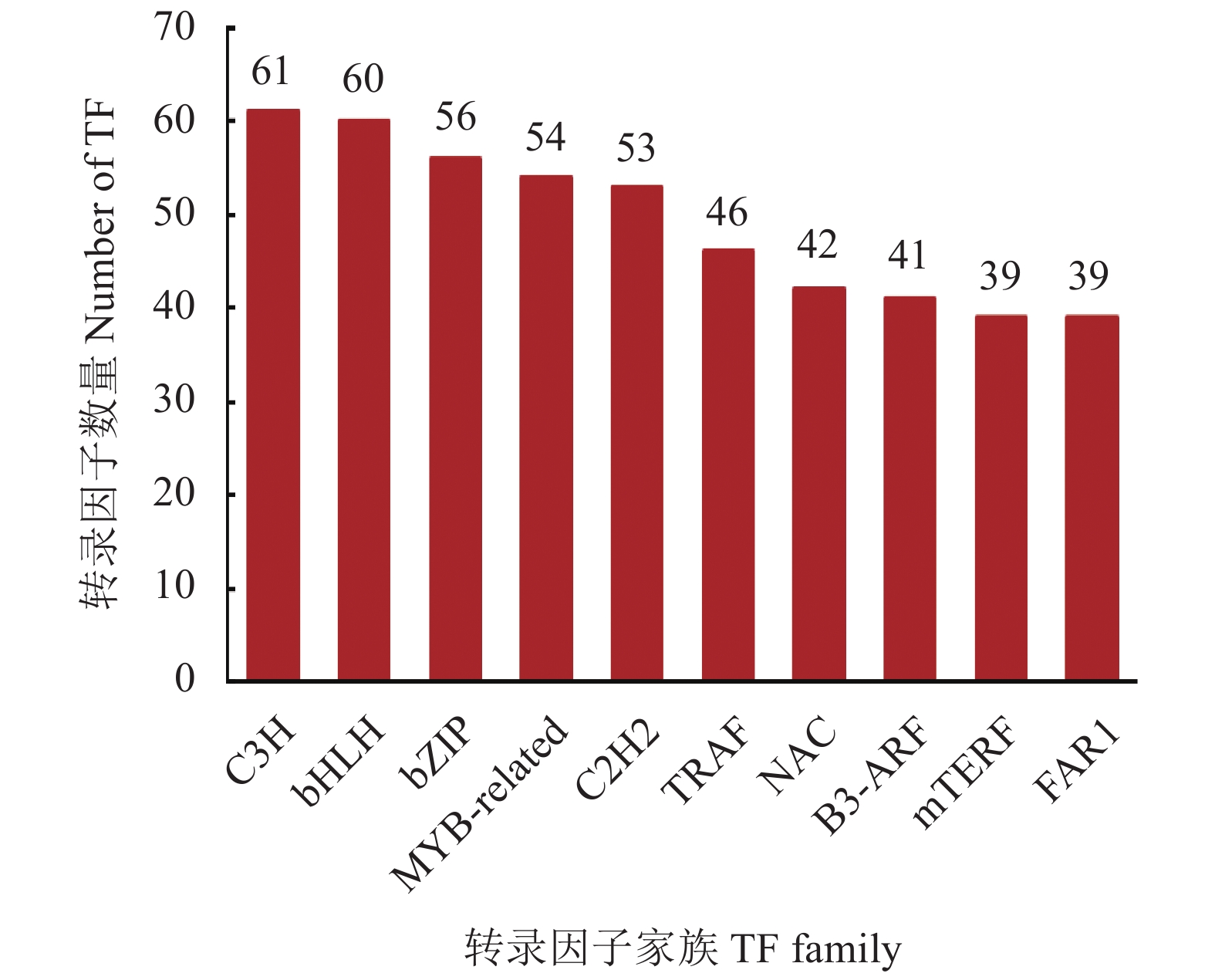
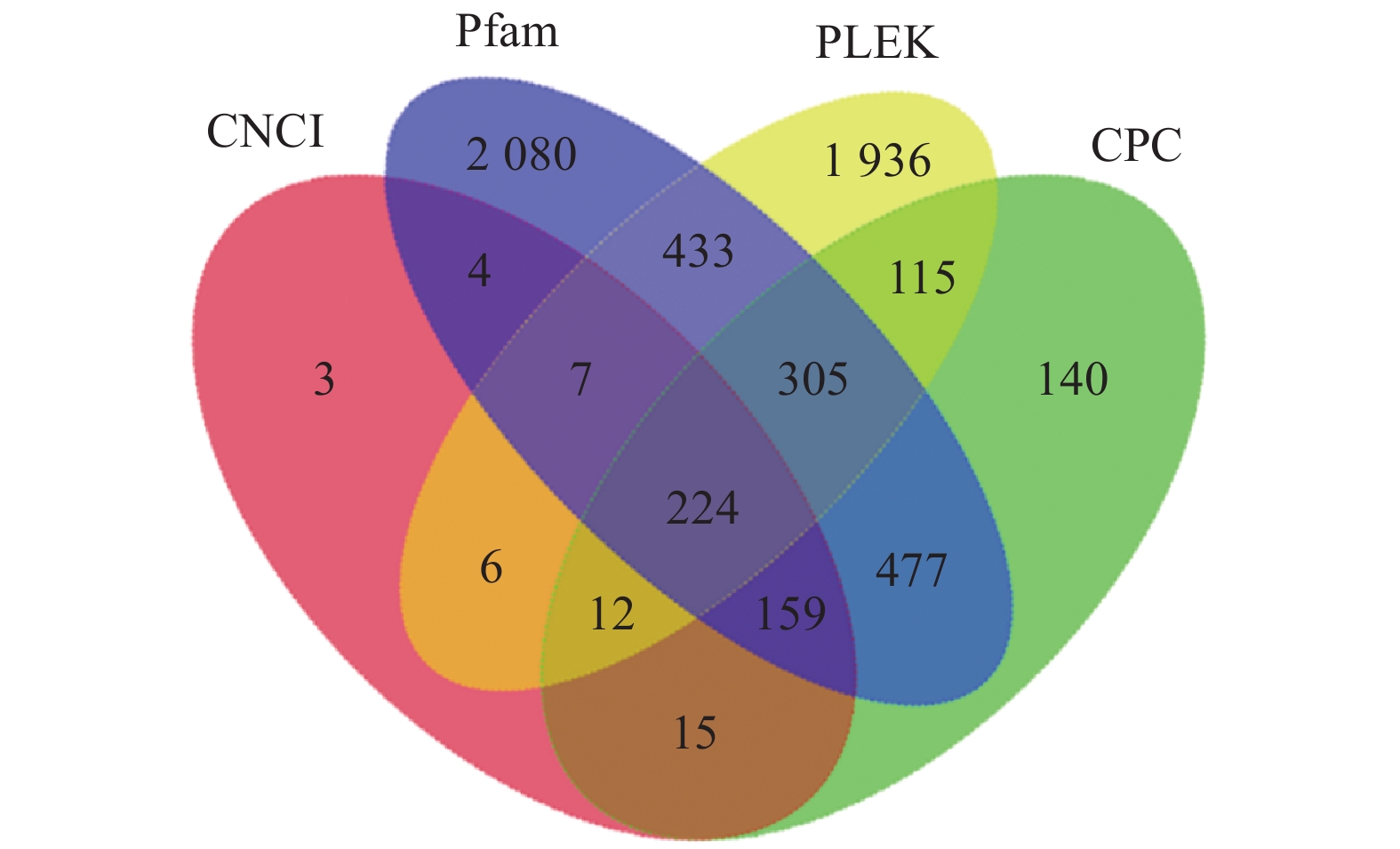
20540 条高质量去冗余转录本。利用NR、GO、NT、Pfam、COG/KOG、 SwissProt、KEGG等7个数据库对高质量去冗余序列进行基因功能注释,共有19909 条转录本至少在1个数据库得到注释,有8888 条转录本在NR、NT、COG/KOG、KEGG、GO等5个公共数据库中均有注释。GO注释结果显示,14911 条转录本共富集在包含生物学过程、细胞组成和分子功能三大类的53个条目中。KEGG分析显示,19701 条转录本序列被富集到6条主通路和44条子通路。COG/KOG注释发现,13204 条转录本被注释,其中注释最多的功能类别为一般功能预测。对转录本结构分析发现,共预测到978个转录因子、224条长非编码RNA、7167 个SSR。对萜类化合物生物合成途径进行挖掘,共鉴定到48个转录本(16个关键酶候选基因)参与萜类骨架生物合成。结论本研究成功对畲药十二时辰基源植物重瓣铁线莲的根进行了全长转录组测序,获得该物种的全长转录组相关基因功能信息,填补了重瓣铁线莲基因信息的空白,为深入研究重瓣铁线莲的调控网络、生物学特征、相关代谢途径、信号通路及分子机制等提供参考。
Abstract:ObjectiveTranscription of herbal plant Clematis florida Thunb. var. plena D. Don was used to identify the key genes involved in the metabolic pathways of the active compounds in the famed “Shiershichen” of She medicine.
MethodPacBio single-molecule real-time (SMRT) was employed to sequence the full-length transcriptome of the roots of the herbal plant that contains the active compounds of the She medicine. Functional annotation, gene structure, and mining of the terpenoid biosynthetic pathway were conducted using bioinformatics tools to secure the transcript data.
ResultFrom the 62.21 G polymerase read bases generated,
20540 non-redundant high-quality transcript sequences were identified after data treatment. At least one of the 7 major databases including NR, NT, Pfam, COG/KOG, SwissProt, GO, and KEGG applied annotated the gene functions of19909 transcripts, and8888 were in NR, NT, COG/KOG, KEGG, and GO. The GO annotation showed14911 transcripts enriched in 53 terms that included biological processes, cellular components, and molecular functions. The KEGG database annotated19701 transcripts and classified them into 6 major pathways and 44 sub-pathways with the largest number of transcripts enriched in metabolic pathways. The COG/KOG annotation identified13204 transcripts with the general function prediction being the most predominant. There were 978 transcription factors, 224 LncRNAs, and7167 SSRs predicted, and 48 transcripts with 16 key candidate genes involved in the terpenoid backbone biosynthesis identified.ConclusionThe full-length transcriptome sequencing on the roots of C. florida was successfully obtained with the comprehensive gene function information. It provided a basis for further studies on the regulatory network, biological characteristics, related metabolic pathways, signaling pathways, and molecular mechanisms associated with the well-known She herbal medicine.
-
Keywords:
- Clematis florida /
- full-length transcriptome /
- transcription /
- LncRNA
-
0. 引言
【研究意义】芦笋(Asparagus officinalis L.)又名石刁柏,是百合科(Liliaceae)天门冬属(Asparagus)多年生宿根植物,具有抗肿瘤和抗氧化等功效,是世界公认的高级保健蔬菜,享有“蔬菜之王”美誉[1−2]。芦笋茎枯病由天门冬拟茎点霉菌[Phomopsis asparagi(Sacc.)Bubak]侵染引起,主要为害芦笋茎秆,在中国、日本、泰国和印尼等亚洲国家芦笋种植区每年均有发生,严重影响芦笋的产量和品质[3−4]。目前,施用杀菌剂是防治芦笋茎枯病的有效手段,但面临着病原菌抗药性、食品安全和环境污染等问题[5]。利用作物品种的抗病性是防治作物病害最经济、有效和安全的措施[6]。芦笋是多年生作物,生产周期长,一次种植可采收10年,建立快速准确的芦笋抗茎枯病鉴定方法,选育抗病品种,对芦笋产业健康发展具有重要意义。【前人研究进展】开展品种抗病性鉴定是选育作物抗病品种的重要一环。我国对水稻、小麦、玉米和大豆等大宗作物的主要病害都建立了稳定可靠的抗病性鉴定方法,形成了国家作物品种抗病性鉴定技术规程[7−8]。在芦笋抗茎枯病的抗性鉴定方面,Sonoda等[9]报道了一种芦笋茎部接种的抗病性鉴定方法,但操作复杂、用时长、受环境条件影响大、重复性差。杨迎青等[10]采用苗期人工接种的方法鉴定了31份芦笋种质资源的抗病性水平,缩短了芦笋抗茎枯病鉴定的时间。阮宏椿等[11]采用苗期人工接种的鉴定方法测定了9个主栽品种对芦笋茎枯病的抗病性。苗期人工接种法具有鉴定结果稳定的优点,但仍需要完成播种、定植等繁杂工作,占用温棚面积大,耗时较长,难以针对大批量品种或育种材料开展抗病性鉴定。【本研究切入点】采用田间自然诱发或苗期人工接种法鉴定芦笋的抗病性,工作量大、耗时长,难以快速从大量种质资源中筛选出抗性材料。芦笋抗茎枯病快速准确鉴定方法还有待深入探讨。【拟解决的关键问题】本研究拟通过评价芦笋茎枯病菌毒素浸种法和浸根法、分生孢子悬浮液浸种法和分生孢子悬浮液灌根法等4种方法与田间自然诱发法对供试芦笋品种抗病性鉴定结果的一致性,建立芦笋抗茎枯病快速准确鉴定的新方法,为芦笋抗病育种和茎枯病的防治提供技术支撑。
1. 材料与方法
1.1 供试材料
供试菌株:供试芦笋茎枯病菌菌株FJ58采集自福建省莆田市涵江区庄边镇吉云村,由福建省农业科学院植物保护研究所分离鉴定,测定致病性并保存。
供试芦笋品种:格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2(感病对照品种),由福建省农业科学院植物保护研究所收集并保存。
供试培养基:马铃薯葡萄糖琼脂培养基(PDA培养基),取200 g马铃薯进行切片,加水煮沸20 min,过滤,上清液加入葡萄糖20 g、琼脂粉16 g,加热至琼脂粉完全溶解,加水定容至
1000 mL,在121 ℃下高压湿热灭菌25 min。马铃薯葡萄糖培养基(PDB培养基),取200 g马铃薯进行切片,加水煮沸20 min,过滤,上清液加入葡萄糖20 g,加水定容至1000 mL,在121 ℃下高压湿热灭菌25 min。1.2 方法
1.2.1 芦笋茎枯病菌孢子悬浮液的制备
芦笋茎枯病菌孢子悬浮液的制备参照杨迎青等[10]的方法 ,并稍作修改。将保存于滤纸片上的芦笋茎枯病菌菌株FJ58转至PDA培养基平板上,28 ℃培养5 d,用直径5 mm的打孔器在菌落边缘打取菌饼,将菌饼转入PDA培养基平板,28 ℃培养14 d,用毛刷和适量无菌水洗下分生孢子器,用灭菌镊子捏碎分生孢子器,释放分生孢子,过滤后血球计数板测定分生孢子浓度,分别配制成1.0×103、1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液,置于4 ℃冰箱保存,备用。
1.2.2 芦笋茎枯病菌毒素的制备
参照李俊萍等[12]的方法,并稍作修改。将芦笋茎枯病菌菌株FJ58转至PDA培养基平板上,28 ℃培养5 d,用直径5 mm的打孔器在菌落边缘打取菌饼,将菌饼转入装有PDB培养基的三角瓶中,28 ℃、150 r·min−1振荡培养7 d,用灭菌滤纸过滤后收集滤液,并用细菌过滤器(0.22 μm)过滤,收集芦笋茎枯病菌粗毒素液。将芦笋茎枯病菌粗毒素液用无菌水稀释,分别配制成芦笋茎枯病菌粗毒素原液∶无菌水(体积比)=1∶0、7∶1、3∶1、1∶1、1∶3和1∶7的稀释液,置于4 ℃冰箱保存备用。
1.2.3 芦笋茎枯病菌毒素浸种法
将供试芦笋种子用10% H2O2浸泡消毒15 min,用无菌水冲洗3次后,然后分别用无菌水浸种22、20、16、8、0 h,再放入芦笋茎枯病菌FJ58的毒素中分别浸泡2、4、8、16、24 h,每个处理的浸种时间均为24 h,同时设无菌水浸种24 h的处理为对照。每处理50粒种子,3次重复。将浸种处理后的种子置于装有灭菌泥炭土的育苗盒中,将育苗盒置于28 ℃、相对湿度90%、光周期12L∶12D的培养箱内培育14 d,调查种子萌发率,计算茎枯病菌毒素对芦笋种子的萌发抑制率。采用DPS 9.5软件Duncan's新复极差法(P < 0.05)进行芦笋种子的萌发抑制率差异显著性分析,并根据试验结果以毒素浸种24 h对芦笋种子的萌发抑制率评价芦笋品种的抗病性[13]。高抗(HR):种子萌发抑制率=0%;抗病(R):0%<种子萌发抑制率≤1%;中抗(MR):1%<种子萌发抑制率≤5%;中感(MS):5%<种子萌发抑制率≤10%;感病(S):10%<种子萌发抑制率≤15%;高感(HS):15%<种子萌发抑制率。
1.2.4 芦笋茎枯病菌毒素浸根法
将供试芦笋种子用10% H2O2浸泡消毒15 min,用无菌水冲洗3次,然后放入无菌水浸泡24 h,置于28 ℃、90%相对湿度的培养箱中催芽3 d,将萌芽的种子置于育苗盒空穴中。每粒芦笋种子分别加入制备的不同芦笋茎枯病毒素稀释液2 mL,以无菌水作对照,每处理30粒种子,3次重复。将育苗盒置于28 ℃、相对湿度90%、光周期12L∶12D的培养箱内培育7 d观察并测量根长,计算不同茎枯病菌毒素稀释液对芦笋根生长的抑制率。采用DPS 9.5软件Duncan's新复极差法(P < 0.05)进行芦笋根生长的抑制率差异显著性分析,并根据试验结果以毒素原液对芦笋根生长的抑制率评价芦笋品种的抗病性。高抗(HR):芦笋根生长的抑制率≤50%;抗病(R):50%<芦笋根生长的抑制率≤60%;中抗(MR):60%<芦笋根生长的抑制率≤70%;中感(MS):70%<芦笋根生长的抑制率≤80%;感病(S):80%<芦笋根生长的抑制率≤90%;高感(HS):90%<芦笋根生长的抑制率。
1.2.5 芦笋茎枯病菌孢子悬浮液浸种法
将供试芦笋种子用10% H2O2浸泡消毒15 min,然后用无菌水冲洗3次,分别放入芦笋茎枯病菌1.0×103、1.0×104、1.0×105个孢子·mL−1分生孢子悬浮液中浸泡15 min,以无菌水浸泡为对照,每处理30粒种子,3次重复。将处理后种子置于装有灭菌泥炭土的育苗盒中,将育苗盒置于28 ℃、90%相对湿度、光周期12L∶12D的培养箱内培育14 d,调查种子萌发率。计算茎枯病菌对芦笋种子的萌发抑制率。采用DPS 9.5软件Duncan's新复极差法(P < 0.05)进行芦笋种子的萌发抑制率差异显著性分析,并根据试验结果以1.0×105个孢子·mL−1的分生孢子悬浮液浸种对种子萌发的抑制率评价芦笋品种的抗病性。高抗(HR):种子萌发抑制率=0%;抗病(R):0%<种子萌发抑制率≤20%;中抗(MR):20%<种子萌发抑制率≤40%;中感(MS):40%<种子萌发抑制率≤60%;感病(S):60%<种子萌发抑制率≤80%;高感(HS):80%<种子萌发抑制率。
1.2.6 芦笋茎枯病菌孢子悬浮液灌根法
将供试芦笋种子用10% H2O2浸泡消毒15 min,用无菌水冲洗3次,然后放入无菌水浸泡24 h,置于28 ℃、90%相对湿度的培养箱中催芽3 d,将处理后的种子置于装有灭菌泥炭土的育苗盒中,将育苗盒置于28 ℃、相对湿度90%、光周期12L∶12D的培养箱内培育14 d,分别用芦笋茎枯病菌1.0×103、1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液灌根接种,每株芦笋幼苗接种2 mL,以无菌水灌根为对照,每处理30株幼苗,3次重复。灌根处理3 d后逐日观察记载各处理的发病株数,计算发病率。采用DPS 9.5软件Duncan's新复极差法(P < 0.05)进行发病率差异显著性分析,并根据试验结果以1.0×103个孢子·mL−1分生孢子悬浮液灌根的发病率评价芦笋品种的抗病性。高抗(HR):发病率=0%;抗病(R):0%<发病率≤5%;中抗(MR):5%<发病率≤15%;中感(MS):15%<发病率≤30%;感病(S):30%<发病率≤45%;高感(HS):45%<发病率。
1.2.7 田间自然诱发法
将供试芦笋品种60 d苗龄的植株移栽种植于常年发生茎枯病的病圃中,60 d后五点取样,每点随机取10丛,观察记载各供试品种植株发病的茎秆数和调查总茎秆数,计算发病率。采用DPS 9.5软件Duncan's新复极差法(P < 0.05)进行发病率差异显著性分析,以发病率评价芦笋品种的抗病性,评价标准同1.2.6。
1.2.8 数据处理
按下列公式分别计算种子萌发率、种子萌发抑制率、根长抑制率和发病率。
种子萌发率/%=萌发的种子数/种子总数×100 (1) 种子萌发抑制率/%=(对照萌发率−处理萌发率)/对照萌发率×100 (2) 根长抑制率/%=(对照根长−处理根长)/对照根长×100 (3) 发病率/%=发病茎秆数/调查总茎秆数×100 (4) 采用SPSS 19.0分析各方法鉴定结果的相关性。
2. 结果与分析
2.1 芦笋茎枯病菌毒素浸种法
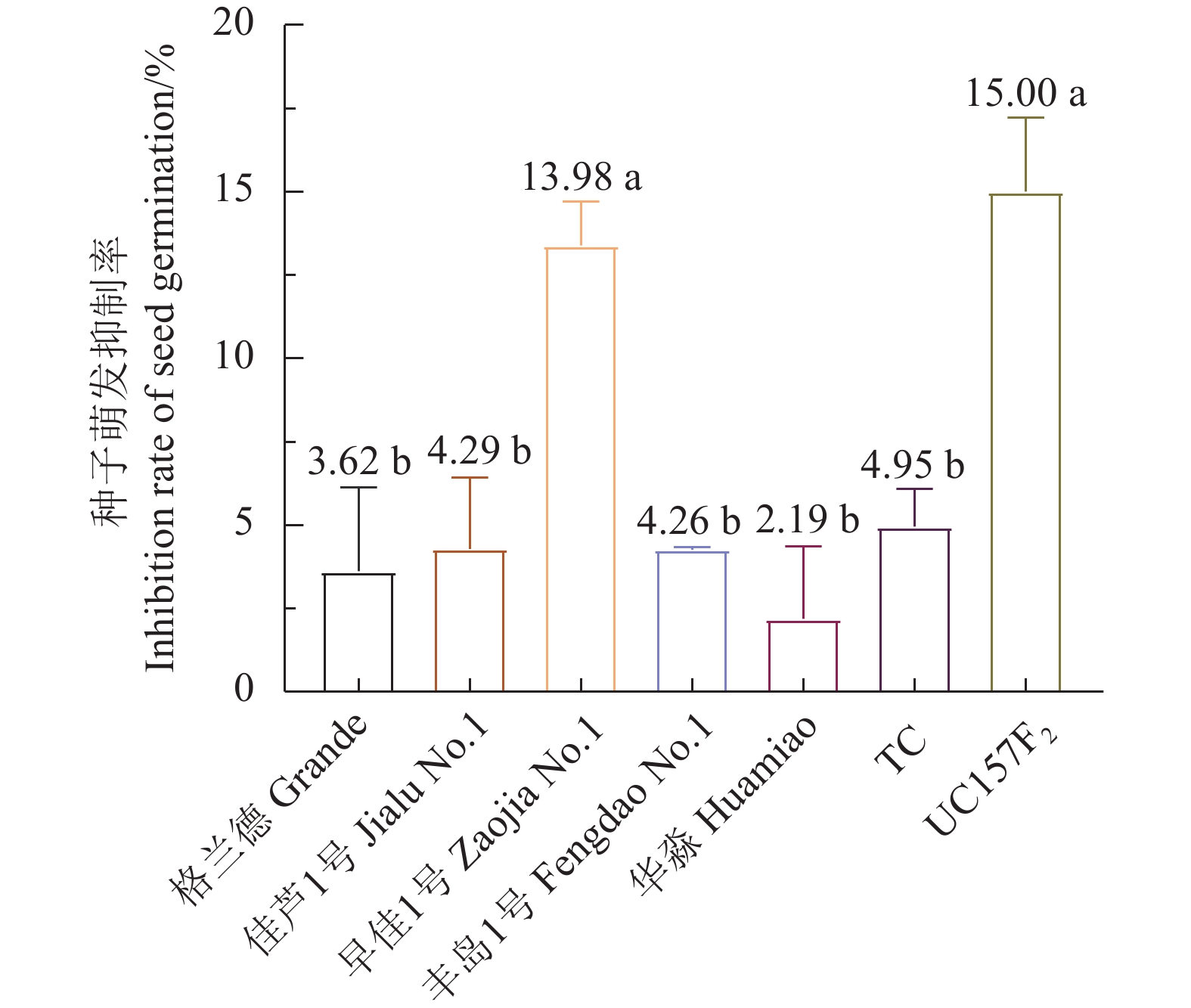
华淼、格兰德、佳芦1号和丰岛1号用芦笋茎枯病菌毒素原液浸种2、4、8、16、24 h的种子萌发率与对照差异不显著;早佳1号和UC157F2种子萌发率与芦笋茎枯病菌毒素浸种时间成反比,其中浸种4、8、16、24 h的萌发率显著低于对照;TC用茎枯病菌毒素浸种24 h的萌发率显著低于对照(表1)。芦笋茎枯病菌毒素浸种24 h对格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2种子萌发抑制率分别为3.62%、4.29%、13.98%、4.26%、2.19%、4.95%和15.00%,对早佳1号和UC157F2种子萌发抑制率显著高于其他供试品种。供试的7个品种被划分为感病和中抗2个类型,感病品种为UC157F2和早佳1号;中抗品种为格兰德、佳芦1号、丰岛1号、华淼和TC(图1)。
表 1 芦笋茎枯病菌毒素不同浸种时间对芦笋种子萌发率的影响Table 1. Effect of soaking time in mycotoxin-containing solution on asparagus seed germination rate浸种时间
Soaking time/h种子萌发率
Germination rate of seed /%格兰德
Grande佳芦1号
Jialu No.1早佳1号
Zaojia No.1丰岛1号
Fengdao No.1华淼
HuamiaoTC UC157F2 2 90.67a 92.00a 94.00a 92.00a 90.00a 92.67ab 90.67ab 4 91.33a 92.00a 91.33b 92.00a 90.67a 90.67ab 88.00bc 8 90.00a 91.33a 89.33b 92.00a 89.33a 92.00ab 85.33cd 16 88.00a 90.67a 84.67c 89.33a 90.67a 92.67ab 82.67de 24 89.33a 89.33a 82.00d 90.00a 89.33a 89.33b 79.33e 0 (CK) 92.00a 93.33a 94.67a 94.00a 91.33a 94.00a 93.33a 同列数据后不同小写字母表示差异显著(P<0.05)。下同。
Data with different lowercase letters on a column indicate significant differences at P<0.05. Same for below.![]() 图 1 芦笋茎枯病菌毒素浸种24 h对芦笋种子萌发的抑制率不同小写字母表示经Duncan氏新复极差法检验差异显著(P < 0.05)。下同。Figure 1. Germination inhibition rate of asparagus seeds soaked in mycotoxin-containing solution for 24 hThose with different lowercase letters indicate significant difference at P<0.05 by Duncan’s new multiple range test. Same for below.
图 1 芦笋茎枯病菌毒素浸种24 h对芦笋种子萌发的抑制率不同小写字母表示经Duncan氏新复极差法检验差异显著(P < 0.05)。下同。Figure 1. Germination inhibition rate of asparagus seeds soaked in mycotoxin-containing solution for 24 hThose with different lowercase letters indicate significant difference at P<0.05 by Duncan’s new multiple range test. Same for below.2.2 芦笋茎枯病菌毒素浸根法
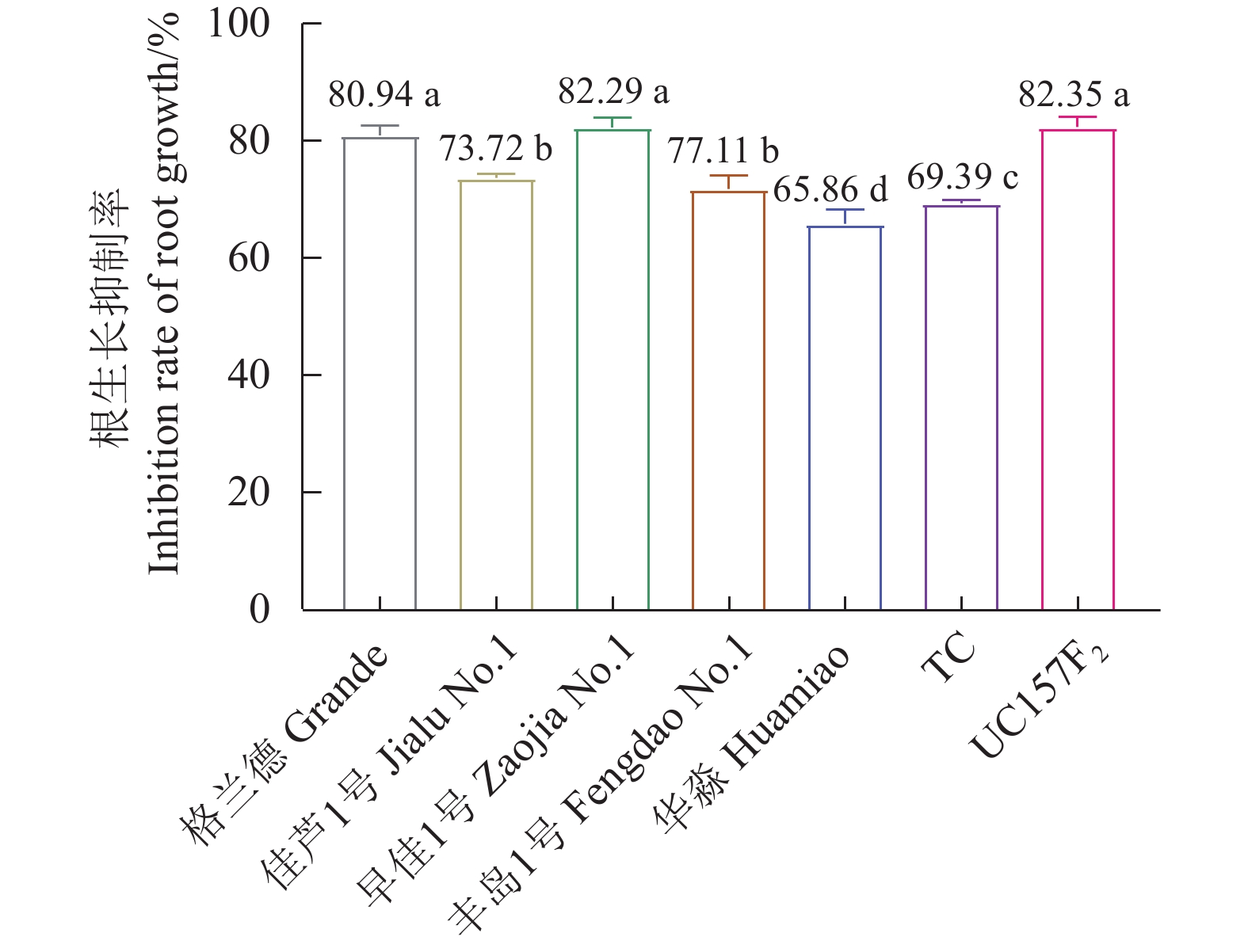
芦笋茎枯病菌毒素浸根处理对芦笋根生长的抑制率与毒素浓度成正比,粗毒素原液∶无菌水(体积比)=1∶0、7∶1、3∶1、1∶1、1∶3和1∶7等6个处理对芦笋根生长抑制率差异达显著水平,其中毒素∶无菌水=1∶0浸根处理对芦笋根生长抑制率显著强于其他处理(表2)。毒素∶无菌水=1∶0浸根处理对格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2的根生长抑制率分别为80.94%、73.72%、82.29%、71.77%、65.86%、69.39%和82.35%,对华淼和TC根生长抑制率显著低于其他供试品种,而对UC157F2、格兰德和早佳1号的根生长抑制率则显著高于其他品种。供试的7个品种被划分为感病、中感和中抗3个类型,感病品种为UC157F2、早佳1号和格兰德;中感品种为佳芦1号和丰岛1号;中抗品种为TC和华淼(图2)。
表 2 不同浓度芦笋茎枯病菌毒素浸根对芦笋根生长的影响Table 2. Effect of soaking in solutions containing varied concentrations of mycotoxin on growth of asparagus roots粗毒素原液与水体积比
Volume ratio of toxin to water根生长抑制率
The inhibition rate of root growth/%格兰德
Grande佳芦1号
Jialu No.1早佳1号
Zaojia No.1丰岛1号
Fengdao No.1华淼
HuamiaoTC UC157F2 1∶0 80.94a 73.72a 82.29a 71.77a 65.86a 69.39a 82.35a 7∶1 74.96b 68.58b 77.12b 67.99b 62.30b 64.60b 77.78b 3∶1 68.99c 64.56c 72.83c 63.02c 58.45c 60.95c 74.75c 1∶1 65.45d 60.85d 68.32d 58.61d 54.54d 55.60d 70.22d 1∶3 60.82e 57.12e 63.80e 53.59e 49.11e 50.54e 66.54e 1∶7 57.28f 55.10f 59.58f 46.00f 43.44f 46.61f 60.77f 2.3 芦笋茎枯病菌分生孢子悬浮液浸种法
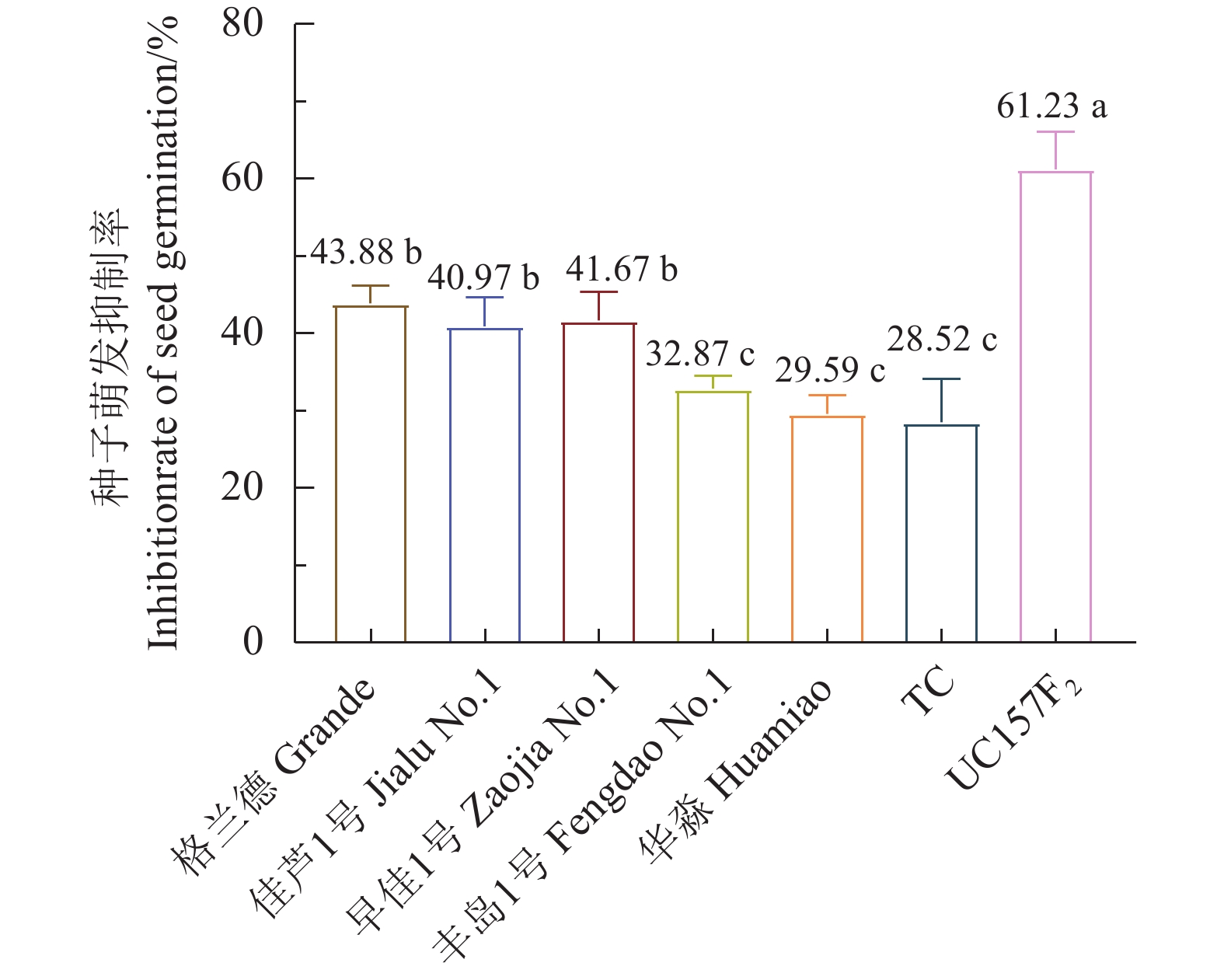
芦笋茎枯病菌1.0×103、1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理对供试7个芦笋品种的种子萌发均有较强的抑制作用,种子萌发率与对照相比均达显著水平。1.0×103、1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理后格兰德和华淼种子萌发率差异均达显著水平。1.0×103、1.0×104个孢子·mL−1的分生孢子悬浮液浸种处理后对佳芦1号、早佳1号、丰岛1号和UC157F2的种子萌发率影响均不显著;1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理对这4个品种种子萌发率差异影响也均不显著,而1.0×103、1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理对这4个品种种子萌发率差异均达显著水平。1.0×103、1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理对TC的种子萌发影响差异不显著。1.0×105个孢子·mL−1的分生孢子悬浮液浸种处理对格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2的种子萌发抑制率分别为:43.88%、40.97%、41.67%、32.87%、29.59%、28.52%和61.23%,供试的7个品种被分为感病、中感和中抗3个类型,感病品种为UC157F2;中感品种为格兰德、佳芦1号和早佳1号;中抗品种为TC、华淼和丰岛1号(表3、图3)。
表 3 不同浓度芦笋茎枯病菌分生孢子悬浮液浸种对芦笋种子萌发率的影响Table 3. Effect on germination rate of seeds soaked in varied P. asparagi spore suspensions分生孢子
悬浮液浓度
Concentration of
spore suspension
/(孢子·mL-1)种子萌发率
Germination rate of seed/%格兰德
Grande佳芦1号
Jialu
No.1早佳1号
Zaojia
No.1丰岛1号
Fengdao
No.1华淼
HuamiaoTC UC-
157F21.0×103 68.00b 68.00b 66.00b 73.33b 77.33b 80.00b 49.33b 1.0×104 60.67c 61.33bc 60.67bc 69.33bc 72.00c 72.00b 44.00bc 1.0×105 52.00d 56.67c 56.00c 62.67c 66.67d 68.67b 36.67c 0 (CK) 92.67a 96.00a 96.00a 93.33a 94.67a 96.00a 94.67a 2.4 芦笋茎枯病菌分生孢子悬浮液灌根法
用芦笋茎枯病菌1.0×103个孢子·mL−1的分生孢子悬浮液灌根处理,格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2的发病率分别为87.33%、84.67%、89.33%、88.00%、83.33%、82.00%和86.00%;用1.0×104、1.0×105个孢子·mL−1的分生孢子悬浮液灌根处理,供试芦笋品种发病率均达100%,不能有效区分供试品种的抗病性(图4)。
![]() 图 4 芦笋茎枯病菌1.0×103个孢子·mL−1孢子悬浮液灌根处理的发病结果A:TC对照;B:TC分生孢子悬浮液灌根;C:UC157F2对照;D:UC157F2分生孢子悬浮液灌根。Figure 4. Disease incident of asparagus plants with roots irrigated with 1.0×103P. asparagi spores·mL−1 suspensionA: TC control; B: TC roots irrigated with P. asparagi spore suspension; C: UC157F2 control; D: UC157F2 roots irrigated with P. asparagi spore suspension.
图 4 芦笋茎枯病菌1.0×103个孢子·mL−1孢子悬浮液灌根处理的发病结果A:TC对照;B:TC分生孢子悬浮液灌根;C:UC157F2对照;D:UC157F2分生孢子悬浮液灌根。Figure 4. Disease incident of asparagus plants with roots irrigated with 1.0×103P. asparagi spores·mL−1 suspensionA: TC control; B: TC roots irrigated with P. asparagi spore suspension; C: UC157F2 control; D: UC157F2 roots irrigated with P. asparagi spore suspension.2.5 田间自然诱发鉴定法
在田间自然诱发的情况下,供试品种格兰德、佳芦1号、早佳1号、丰岛1号、华淼、TC和UC157F2的发病率分别为18.83%、16.97%、20.57%、15.47%、14.93%、13.53%和32.93%。UC157F2最感病,发病率显著高于其他品种,其后依次为早佳1号、格兰德、佳芦1号、丰岛1号、华淼和TC。根据品种发病率差异水平,将供试的7个品种划分为感病、中感和中抗3个类型,感病品种为UC157F2;中感品种为早佳1号、格兰德、佳芦1号和丰岛1号;中抗品种为华淼和TC(图5)。
2.6 不同鉴定方法相关性分析
芦笋茎枯病菌毒素浸种法、毒素浸根法和分生孢子悬浮液浸种法均与田间自然诱发法在0.01水平上显著相关,其中分生孢子悬浮液浸种法与田间自然诱发法的相关系数最大,为0.761。孢子悬浮液灌根法与自然诱发法在0.05水平上显著相关,相关系数为0.540;孢子悬浮液灌根法与毒素浸种法、毒素浸根法、孢子悬浮液浸种法的相关性均不显著。芦笋茎枯病菌毒素浸种法、毒素浸根法和孢子悬浮液浸种法的鉴定结果均在0.01水平上显著相关(表4)。
表 4 不同鉴定方法的相关性Table 4. Correlation between evaluation methods for disease resistance of asparagus抗性鉴定方法
Resistance-identification
methods毒素浸种法
Soaking seed
with mycotoxin毒素浸根法
Soaking root
with mycotoxin分生孢子悬浮液浸种法
Soaking seed
with spore suspension分生孢子悬浮液灌根法
Irrigating root
with spore suspension自然诱发法
Field natural
induction method毒素浸种法
Soaking seed with mycotoxin1 0.768** 0.911** 0.267 0.703** 毒素浸根法
Soaking root with mycotoxin0.768** 1 0.682** 0.327 0.682** 分生孢子悬浮液浸种法
Soaking seed with spore suspension0.911** 0.682** 1 0.373 0.761** 分生孢子悬浮液灌根法
Irrigating root with spore suspension0.267 0.327 0.373 1 0.540* 田间自然诱发法
Field natural induction method0.703** 0.682** 0.761** 0.540* 1 **表示在0.01水平(双侧)上显著相关;*表示在0.05水平(双侧)上显著相关。
** indicates significant correlation at P<0.01 (bilateral); * indicates significant correlation at P<0.05 (bilateral).3. 讨论与结论
芦笋茎枯病是芦笋生产上最主要的病害,造成芦笋产量和品质下降[14]。利用抗病品种是防治芦笋茎枯病的重要措施,而在抗病品种培育中,育种亲本、后代群体与新品种(系)的抗病性评判都依赖抗病性鉴定[15]。目前,国内外对芦笋茎枯病的抗病性缺乏系统性、持续性的研究。芦笋抗病性鉴定通常采用田间病圃自然诱发的方法,而田间温度、降雨、湿度等自然条件的变化会直接影响鉴定结果的准确性,且试验用地大、鉴定周期长、工作量大。因此,建立快速准确鉴定芦笋抗茎枯病的方法对筛选和利用抗性材料,培育抗病品种,有效防控芦笋茎枯病具有重要意义。
芦笋茎枯病菌毒素浸种法对芦笋种子的萌发率和出苗率有一定的影响,其中对早佳1号和UC157F2的种子萌发抑制率有较显著影响,对其他供试品种的种子萌发抑制率均在5%以下,这可能与芦笋种子有坚硬皮壳包裹,毒素难以接触胚芽有关。孙丽萍等[13]研究结果也表明,毒素浸种法不能有效鉴定烟草种质对赤星病的抗性。 丁燕芳等[16]研究表明用毒素浸种法鉴定烟草种质对黑胫病抗性简单灵活,可进行多批量的初期筛选,但需注意因种子质量问题(低发芽率)造成“假感病”现象,影响了毒素浸种法的判定效果。本研究采用病菌毒素对种子萌发抑制率评价供试品种的抗病性,消除了因种子质量差造成的“假感病”现象,但仍需注意种子皮壳厚度及坚硬程度对毒素浸种鉴定的影响,选择适宜的浸种时间。
毒素浸根法与田间病圃自然诱发法相比具有简单易行和不易受环境影响等特点[17]。本研究发现,芦笋根生长的抑制率与芦笋茎枯病菌毒素浓度成正比,毒素原液对所有供试芦笋品种根的生长抑制率均在65%以上,对有效区分品种的抗感性有一定的影响。贺红等[17]采用0.6×108 cfu·mL−1 的青枯菌液制备的毒素浸根处理广藿香植株,可较快表现出青枯病的症状,缩短抗病鉴定的过程。王铭等[18]研究表明毒素浸根法鉴定棉花抗黄萎病时最适毒素浓度为15 μg·mL−1,在此浓度下,毒素浸根法的鉴定结果与田间病圃法鉴定结果较一致。在烟草黑胫病抗性鉴定中,毒素浸根法可以初步鉴别抗、感病品种,但是存在不能区分中感和中抗烟草品种的缺陷[16]。棉花黄萎病菌和烟草青枯病菌都侵染寄主的维管束,病菌毒素通常具有较强的致病或致枯作用,病菌毒素处理后寄主快速表现病害症状。可见,毒素浸根法对病菌或毒素造成系统性病害的作物抗性鉴定比较适用,而毒素浓度是影响鉴定结果的关键因素。
本研究采用分生孢子悬浮液灌根处理,1.0×103个孢子·mL−1的分生孢子悬浮液处理下供试芦笋品种的发病率均在80%以上,而1.0×104个孢子·mL−1的分生孢子悬浮液处理下供试芦笋品种的发病率达100%,这种结果不能有效区分供试品种对茎枯病的抗病性。这与芦笋茎枯病菌具有较强的致病性有关,分生孢子悬浮液灌根后病菌孢子快速萌发侵染芦笋幼嫩的根和茎,迅速造成枯萎。周淼平等[19]采用纹枯病菌菌丝体悬浮液浸种小麦种子法测定了小麦对纹枯病的抗病性,鉴定结果与田间幼苗纹枯病抗性鉴定结果有较高的相关性,且操作简便,重复性好。兰海燕[20]认为通过向日葵种子与菌核共培养,根据种子皮壳对病原菌的抗病能力鉴定向日葵对菌核病抗性的方法准确率高,且在播种前即可在室内快速大量进行定向选择,该鉴定方法的生化依据是抗病种子的皮壳上酚的含量比不抗病的高2~3倍。本研究中,芦笋茎枯病菌分生孢子悬浮液浸种法与田间自然诱发法的抗病性鉴定结果相关系数为0.761,一致性较高,可以与田间自然诱发法相结合用于快速筛选芦笋抗茎枯病种质资源。
-
表 1 单分子实时测序数据统计
Table 1 Statistics of sequencing data by SMRT
类别
Type总数
Total number最小长度
Min length/bp最大长度
Max length/bp平均长度
Mean length/bpN50长度
N50 length/bp子序列
Subreads27863322 — — 2233 2469 环形一致性序列
Circular consensus sequence (CCS)595827 54 14994 2488 2659 全长非嵌序列
Full-length non-chimeric read (FLNC)494732 57 14860 2333 2531 优化一致性序列
Polished consensus sequence41768 87 6251 2272 2468 去冗余高质量序列
Isoform20540 87 6251 2248 2457 表 2 转录本KEGG注释结果
Table 2 KEGG annotated transcripts
KEGG主通路
Main pathwayKEGG子通路
Sub-pathway of KEGG转录本数量
Number of
transcriptsKEGG主通路
Main pathwayKEGG子通路
Sub-pathway of KEGG转录本数量
Number of
transcripts细胞过程
Cellular process运输与催化
Transport and catabolism595 代谢
Metabolism异种生物降解和代谢
Xenobiotics biodegradation and metabolism85 细胞群落-原核生物
Cellular community-prokaryotes93 核酸代谢
Nucleotide metabolism198 细胞群落-真核生物
Cellular community-eukaryotes124 萜类和聚酮类代谢
Metabolism of terpenoids and polyketides203 细胞运动
Cell motility39 其他氨基酸代谢
Metabolism of other amino acids333 细胞生长和与死亡
Cell growth and death276 辅助因子和维生素代谢
Metabolism of cofactors and vitamins345 环境信息处理
Environmental
information processing信号转导
Signal transduction984 脂类代谢
Lipid metabolism437 膜运输
Membrane transport51 聚糖生物合成和代谢
Glycan biosynthesis and metabolism171 遗传信息处理
Genetic information processing翻译
Translation806 全局和总览图
Global and overview maps718 转录
Trancription480 能量代谢
Energy metabolism540 复制与修复
Replication and repair176 碳水化合物代谢
Carbohydrate metabolism1010 折叠,分类和降解
Folding, sorting and degradation657 其他生长代谢物合成
Biosynthesis of other secondary metabolites243 人类疾病
Human diseases药物依赖
Substance dependence44 氨基酸代谢
Amino acid metabolism561 神经退行性疾病
Neurodegenerative diseases165 组织系统
Organismal systems感官系统
Sensory system15 感染疾病-病毒
Infectious diseases:Viral496 神经系统
Nervous system294 感染疾病-寄生生物
Infectious diseases:Parasitic217 免疫系统
Immune system359 感染疾病-细菌
Infectious diseases:Bacterial309 分泌系统
Excretory system51 免疫疾病
Immune diseases35 环境适应
Environmental adaptation362 内分泌与代谢疾病
Endocrine and metabolic diseases141 内分泌系统
Endocrine system404 耐药性:抗肿瘤药
Drug resistance: Antineoplastic94 消化系统
Digestive system77 心血管疾病
Cardiovascular diseases88 发育
Development17 癌症:特殊类型
Cancers: Specific types86 循环系统
Circulatory system57 癌症:总览
Cancers: Overview384 老龄化
Aging148 表 3 重瓣铁线莲转录组中与萜类化合物合成相关的转录本
Table 3 Isoforms encoding key enzymes involved in terpenoid biosynthesis in C. florida
合成途径
Pathway关键酶名称
Key enzyme name酶的系统编号
Enzyme No.转录本数
No. of isoformMVA途径
MVA Pathway羟甲基戊二酰辅酶A合酶
hydroxymethylglutaryl-CoA synthase (HMGS)EC:2.3.3.10 1 羟甲基戊二酰辅酶A还原酶
hydroxymethylglutaryl-CoA reductase(HMGR)EC:1.1.1.34 2 甲羟戊酸激酶
mevalonate kinase(MK)EC:2.7.1.36 1 磷酸甲羟戊酸激酶
phosphomevalonate kinase(PMK)EC:2.7.4.2 1 二磷酸甲羟戊酸脱羧酶
diphosphomevalonate decarboxylase (MVD)EC:4.1.1.33 2 MEP途径
MEP Pathway1-脱氧-D-木酮糖5-磷酸合酶
1-deoxy-D-xylulose-5-phosphate synthase(DXS)EC:2.2.1.7 11 1-脱氧-D-木酮糖5-磷酸还原异构酶
1-deoxy-D-xylulose-5-phosphate reductoisomerase(DXR)EC:1.1.1.267 5 2-C-甲基-D-赤藓醇-4-磷酸胱氨酰转移酶
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase(MCT)EC:2.7.7.60 1 4-二磷酸二胺-2-C-甲基-D-赤藓糖醇激酶
4-diphosphocytidyl-2-C-methyl-D-erythritol kinase(CMK)EC:2.7.1.148 1 4-羟基-3-甲丁-2-烯基二磷酸合酶
4-hydroxy-3-methylbut-2-enyl-diphosphate synthase(HDS)EC:1.17.7.1 9 4-羟基-3-甲丁-2-烯基二磷酸还原酶
4-hydroxy-3-methylbut-2-enyl diphosphate reductase(HDR)EC:1.17.7.4 2 分支点
Branch points异戊烯二磷酸Delta-异构酶
Isopentenyl-diphosphate delta-isomerase(IPPI)EC:5.3.3.2 2 牻牛儿基牻牛儿基焦磷酸合酶
geranylgeranyl diphosphate synthase(GGPS)EC: 2.5.1.29 6 法呢基焦磷酸合酶
farnesyl diphosphate synthase(FPPS)EC:2.5.1.10 1 蛋白香叶烯基转移酶 protein farnesyltransferase subunit (FNTB) EC:2.5.1.58 1 内肽酶 endopeptidase(STE24) EC:3.4.24.84 2 -
[1] 黄泽豪, 张月玲. 畲药十二时辰的本草考证 [J]. 中国民族民间医药, 2014, 23(1):2−3,5. HUANG Z H, ZHANG Y L. Textual research on she medicine’s twelve o’clock materia Medica [J]. Chinese Journal of Ethnomedicine and Ethnopharmacy, 2014, 23(1): 2−3,5. (in Chinese)
[2] 何舒澜, 杨谨, 黄娴, 等. 重瓣铁线莲鲨烯合酶SQS基因的克隆及表达分析 [J]. 福建农业学报, 2022, 37(11):1430−1437. HE S L, YANG J, HUANG X, et al. Cloning and expression of squalene synthase gene from Clematis florida Thunb. var. plena D. don [J]. Fujian Journal of Agricultural Sciences, 2022, 37(11): 1430−1437. (in Chinese)
[3] 沈廷明, 黄春情, 吴军军, 等. GC-MS法分析畲药十二时辰花中挥发油成分 [J]. 中草药, 2020, 51(24):6362−6366. SHEN T M, HUANG C Q, WU J J, et al. GC-MS analysis of volatile oil from traditional She medicine Clematis florida var. plena flower [J]. Chinese Traditional and Herbal Drugs, 2020, 51(24): 6362−6366. (in Chinese)
[4] YANG N N, ZHANG Y F, ZHANG H T, et al. The in vitro and in vivo anti-inflammatory activities of triterpene saponins from Clematis florida [J]. Natural Product Research, 2021, 35(24): 6180−6183. DOI: 10.1080/14786419.2020.1833203
[5] SUN K H, MA X H, ZENG X M, et al. A new indole-type alkaloid from the roots of Clematis florida var. plena [J]. Natural Product Research, 2019, 33(20): 2925−2931. DOI: 10.1080/14786419.2018.1510396
[6] WU Y, ZHANG Y F, ZHANG H T, et al. A new cyclopeptide alkaloid from Clematis Florida [J]. Natural Product Research, 2022, 36(7): 1693−1699. DOI: 10.1080/14786419.2020.1809399
[7] 黄丽容, 郭晓云, 胡营, 等. 香附子全长转录组测序及生物信息学分析 [J]. 中国现代中药, 2023, 25(7):1428−1440. HUANG L R, GUO X Y, HU Y, et al. Full-length transcriptome sequencing and bioinformatics analysis of Cyperus rotundus L [J]. Modern Chinese Medicine, 2023, 25(7): 1428−1440. (in Chinese)
[8] 米琪, 赵艳丽, 徐萍, 等. 滇黄精全长转录组测序及生物信息学分析 [J]. 药学学报, 2024, 59(6):1864−1872. MI Q, ZHAO Y L, XU P, et al. Full-length transcriptome sequencing and bioinformatics analysis of Polygonatum kingianum [J]. Acta Pharmaceutica Sinica, 2024, 59(6): 1864−1872. (in Chinese)
[9] HE B, SHAN T Y, XU J Y, et al. Full-length transcriptome profiling of Acanthopanax gracilistylus provides new insight into the kaurenoic acid biosynthesis pathway [J]. Physiology and Molecular Biology of Plants, 2024, 30(3): 383−399. DOI: 10.1007/s12298-024-01436-7
[10] BUCHFINK B, XIE C, HUSON D H. Fast and sensitive protein alignment using DIAMOND [J]. Nature Methods, 2015, 12(1): 59−60. DOI: 10.1038/nmeth.3176
[11] ZHENG Y, JIAO C, SUN H H, et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases [J]. Molecular Plant, 2016, 9(12): 1667−1670. DOI: 10.1016/j.molp.2016.09.014
[12] THIEL T, MICHALEK W, VARSHNEY R K, et al. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L. ) [J]. TAG Theoretical and Applied Genetics Theoretische und Angewandte Genetik, 2003, 106(3): 411−422. DOI: 10.1007/s00122-002-1031-0
[13] SUN L, LUO H T, BU D C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts [J]. Nucleic Acids Research, 2013, 41(17): e166. DOI: 10.1093/nar/gkt646
[14] KANG Y J, YANG D C, KONG L, et al. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features [J]. Nucleic Acids Research, 2017, 45(W1): W12−W16. DOI: 10.1093/nar/gkx428
[15] FINN R D, COGGILL P, EBERHARDT R Y, et al. The pfam protein families database: Towards a more sustainable future [J]. Nucleic Acids Research, 2016, 44(D1): D279−D285. DOI: 10.1093/nar/gkv1344
[16] LI A M, ZHANG J Y, ZHOU Z Y. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme [J]. BMC Bioinformatics, 2014, 15(1): 311. DOI: 10.1186/1471-2105-15-311
[17] 沈廷明, 陈懿冲, 黄春情, 等. 畲药十二时辰的质量标准研究 [J]. 海峡药学, 2023, 35(8):36−41. SHEN T M, CHEN Y C, HUANG C Q, et al. Quality standard of she medicine Clematis florida var. plena [J]. Strait Pharmaceutical Journal, 2023, 35(8): 36−41. (in Chinese)
[18] 陈懿冲, 沈廷明, 陈艳柠, 等. 基于网络药理学探讨畲药十二时辰镇痛作用机制 [J]. 海峡药学, 2024, 36(1):1−6. CHEN Y C, SHEN T M, CHEN Y N, et al. Based on the network pharmacology to explore the mechanism of analgesic action of she medicine Clematis florida var. flore-pleno [J]. Strait Pharmaceutical Journal, 2024, 36(1): 1−6. (in Chinese)
[19] JIANG C H, BI Y K, MO J B, et al. Proteome and transcriptome reveal the involvement of heat shock proteins and antioxidant system in thermotolerance of Clematis florida [J]. Scientific Reports, 2020, 10(1): 8883. DOI: 10.1038/s41598-020-65699-2







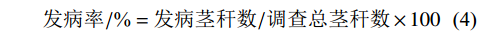
 下载:
下载: