Expressions and Functions of Flowering Locus Ts in Narcissus tazetta var. chinensis Roem
-
摘要:目的 开花基因座T(Flowering Locus T,FT)基因广泛参与植物的生长发育,在调控开花、地下茎发育、种子萌发和逆境胁迫等过程中发挥重要作用。揭示FT基因在中国水仙(Narcissus tazetta var. chinensis Roem)中的表达模式和功能可为中国水仙花期调控提供理论依据。方法 基于中国水仙转录组数据,对FT 基因进行筛选,获得4个中国水仙FT基因。利用荧光定量PCR技术分析它们在中国水仙不同组织和花芽分化不同时期的表达模式,并在拟南芥(Arabidopsis thaliana)中超量表达。利用荧光定量PCR技术分析转NtFT1和NtFT4基因拟南芥中SUPPRESSOR OF OVEREXPRESSION OF CO 1(SOC1)、LEAFY(LFY)和APETALA 1(AP1) 基因表达水平。结果 克隆了4个中国水仙FT同源基因NtFT1、NtFT2、NtFT3和NtFT4,除NtFT3外,中国水仙NtFT都具有FT的保守基序;系统进化分析显示,NtFT1属于FT-like I进化枝,NtFT2、NtFT3和NtFT4同属于FT-like II类进化枝。不同的NtFT基因在中国水仙组织器官和花芽分化不同时期的表达模式存在差异:NtFT1和NtFT3在花中表达量最高,NtFT2在叶片中表达量最高,NtFT4在鳞片中的表达量最高;NtFT1在花芽分化过程中均呈先上升后下降的趋势,NtFT2整个主芽分化过程中变化幅度不大,NtFT3和NtFT4在整个花芽分化期表达量都较低。异位转化拟南芥结果显示,与野生型拟南芥相比,过表达NtFT1和NtFT2的拟南芥提早开花,过表达NtFT3拟南芥开花时间与野生型植株无明显差异,转NtFT4基因的拟南芥植株推迟开花。过表达NtFT1拟南芥中SOC1、LFY和AP1基因表达量上升。结论 在中国水仙中存在多个FT基因,在调控开花的功能上存在差异,NtFT1促进开花,NtFT4抑制开花。Abstract:Objective Expressions and functions of Flowering Locus T (FT), the genes widely involved in plant growth, flowering regulation, root development, and seed germination, in Chinese narcissus were studied.Method Four FTs were identified from the transcriptome data on Narcissus tazetta var. chinensis Roem by bioinformatics analysis. Expressions of the genes in various tissues and flower buds at differentiation stages were determined by fluorescence quantitative PCR and further confirmed by overexpressing it in Arabidopsis thaliana.Result The 4 homologous FTs, i.e., NtFT1, NtFT2, NtFT3, and NtFT4, were cloned using RT-PCR. All of them, except NtFT3, had conserved motifs. Phylogenetically; NtFT1 belonged to the FT-like I branch, while NtFT2, NtFT3, and NtFT4, to the FT-like II branch. In tissues, organs, and flower buds at differentiation stages, they expressed differently. The highest expressions of NtFT1 and NtFT3 were in the flowers, those of NtFT2 in the leaves, and NtFT4 in the scales. At different flower bud differentiation stages, the expressions differed significantly as well, as NtFT1 decreased followed as an increase during differentiation, but NtFT2 changed little throughout. The expressions of NtFT3 and NtFT4 were relatively low in the entire flower bud differentiation period. The ectopic transformation of A. thaliana showed the overexpressed NtFT1 and NtFT2 led to early flowering in comparison to the wild type. But no effect was associated with NtFT3, but delayed flowering was found on the transgenic NtFT4 Arabidopsis plants. In addition, the expressions of SOC1, LFY, and AP1 in the Arabidopsis plants rose with NtFT1 overexpression.Conclusion There were multiple FTs in Chinese narcissus that differed in regulating the flowering—NtFT1 promoted, while NtFT4 inhibited, the process.
-
0. 引言
【研究意义】开花是植物生长发育的重要活动之一,受外界环境和植物内源信号的共同调控。高等植物的开花途径可分为光周期途径、春化途径、赤霉素途径、温度途径、自主途径和年龄途径等6条途径[1],它们相互作用,形成调控网络共同调控植物开花。FT、LFY、SOC1等基因被认为是植物开花途径中的整合基因[2]。FT 基因是Phosphatidylethanolamine-binding protein(PEBP)家族的成员之一[3]。PEBP家族成员广泛存在于植物、动物和微生物中,在植物的生长发育中发挥重要作用[3,4]。在光周期调控途径中,在长日照条件下CONSTANS(CO)诱导下游基因FT表达从而促进植物开花[5]。中国水仙是石蒜科(Amaryllidaceae)水仙属(Narcissus)多年生球根草本植物,多花水仙的变种。中国水仙花芽分化主要受高温的影响[6],乙烯处理可以增加花葶和小花数目[7],但其花芽分化的调控机制仍不清楚。因此对中国水仙FT基因进行研究具有重要意义。【前人研究进展】前人研究结果表明多种植物的FT基因具有促进开花的能力。如野菊花(Chrysanthemum morifolium)的CsFTL1、CsFT2和CsFTL3 [8−11],玫瑰(Rosa rugosa)的RoFT [12],大豆(Glycine max)的GmFT2和GmFT5a [13]等。近年来,在一些植物中发现FT基因也能够抑制开花。如矮牵牛(Petunia hybrida)的PhFT1[14]、龙眼(Dimocarpus longan)的Dl FT1[15]、甘蔗(Saccharum officinarum)的ScFT1[16]等。随着研究的深入,发现高等植物的FT基因除了与植物的开花有关外,还参与调控块茎形成[17]、侧枝发育[18]、气孔开放[19]、种子萌发[20]、植物营养生长[21]和植物的逆境胁迫响应[22]等。【本研究切入点】目前已从中国水仙中克隆了3个FT基因,并研究了它们在中国水仙休眠期的表达模式[23]。但目前只对FT1基因开展了功能分析,发现其能够促进转基因拟南芥提前开花[24],其他FT基因的表达模式和功能研究还未见报道。【拟解决的关键问题】本研究克隆了4个中国水仙FT基因,通过荧光定量PCR技术分析其在不同组织和花芽分化不同发育时期的表达情况,并通过转化拟南芥进行功能分析,以期进一步丰富单子叶植物的FT基因功能研究,并为FT基因的开发利用提供资料。
1. 材料与方法
1.1 试验材料
试验材料为中国水仙金盏银台,分别于2019 年7月8日、7月17日、7月30日、8月7日、8月17日和8月29日 取处于不同分化时期的主芽;12月份取中国水仙的根、鳞茎盘、鳞片、叶和花。所有材料取完后立放入液氮,−80 ℃冰箱保存。
1.2 试验方法
1.2.1 中国水仙 FT基因的克隆及生物信息学分析
根据功能注释检索中国水仙转录组数据库获得FT/TFL1蛋白结构域的所有序列,去除不完整蛋白序列。通过 NCBI CDD search、SMART(http://smart.embl-heidelberg.de/smart/set_mode.cgiNORMAL=1)和 motif (https://meme-suite.org/meme/tools/meme)对检索结果整合筛选,去除不含有FT完整结构域的序列,得到FT基因家族候选成员。根据各基因的序列设计特异引物(表1),以中国水仙花芽cDNA为模板克隆FT基因,分别命名为NtFT1、NtFT2、NtFT3和NtFT4。
表 1 克隆中国水仙NtFT基因引物Table 1. Primers for cloning NtFT of N. tazetta引物名称
Primer name上游引物序列(5′-3′)
Upstream primer sequence (5′-3′)下游引物序列(5′-3′)
Downstream primer sequence (5′-3′)NtFT1 TTTCCGCTTATATCTCTTCTGGGAC TCGGGAAGTAGCAAGACGATCAAAC NtFT2 ATGTTGAGAGAGAGGGTACC TCAGCAAAGTCCTGAGAACCTTCTT NtFT3 GAAGTAGTCATGTTGAGAGAGAGG GTATCACATATTGCATGGCTTAGG NtFT4 GGTTAAGAGACAGAATGCCGAT TTTATGTCATTTATCGTCTG CTAG 在 NCBI 网站上下载拟南芥、玉米(Zea mays)、洋葱(Allium cepa)、水稻(Oryza sativa)和郁金香(Tulipa gesneriana)等植物的FT同源蛋白序列,通过MEGA-X 7.0软件构建进化树,采用邻接法(Neighbor-joining, NJ)构建进化树。
1.2.2 FT基因的表达分析
分别提取中国水仙不同组织和不同分化时期的主芽RNA并逆转录成cDNA。使用翊圣生物公司的 qPCR SYBR试剂盒进行荧光定量PCR,分析各样品的FT基因表达情况,各基因的定量引物见表2。每个样品3次生物学重复,采用2−ΔΔCt法计算相对表达量。
表 2 FT基因表达qRT-PCR引物Table 2. Primers for qRT-PCR of FT gene引物名称
Primer name上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)上游引物序列(5’-3’)
Downstream primer sequence (5′-3′)NtActin GTTGACCCACCACTAAGAACAATG TGCCCAGAAGTGCTATTCCAG QNtFT1 CCAGCCAAAGGTTGAAGTCG CCCTGTGGTTCCTGGTATG QNtFT2 CTTATGAGAGCCCTCGAACACC CGCACACAGTTTGTTGAACTTCC QNtFT3 CGCACACAGTTTGTTGAACTTCC CACTGGTTGGTGACAGACATACC QNtFT4 TGGCAGGATGCGATGCAAGA CACGATGCGATGAATCCCCGA 1.2.3 中国水仙NtFT基因植物表达载体的构建及遗传转化分析
利用In fusion方法构建植物表达载体。用限制性内切酶Hind Ⅲ和EcoR Ⅰ对pSAK277进行双酶切,电泳后回收大片段获得线性化载体。根据线性化载体酶切部位的序列设计特异性引物(表3),进行PCR扩增获得添加特异序列的各目的基因产物。利用In fusion方法将中国水仙各FT基因插入到pSAK277质粒中,构建pSAK277-NtFT1、pSAK277-NtFT2、pSAK277-NtFT3和pSAK277-NtFT4表达载体。采用花序浸染法分别将4个FT基因转化到Col野生型拟南芥中,获得转基因植株。将抗性阳性T0代转基因拟南芥移植到基质中,待其长到4~5片真叶时,提取叶片DNA进行PCR扩增,获得转基因植株。各基因检测的引物如表1所示。
表 3 pSAK277表达载体构建引物Table 3. Primers for constructing pSAK277 expression vector引物名称
Primer name上游引物序列(5′-3′)
Upstream primer sequence (5′-3′)下游引物序列(5′-3′)
Upstream primer sequence (5′-3′)277NtFT1 ACTAGTGGATCCAAAGAATTCATGA
GTAGGGATCCTTTGGTTATTGAGAAGTACTCTCGAGAAGCTT
TTAGGGGTACATCCTCCGGCCACCA277NtFT2 ACTAGTGGATCCAAAGAATTCAGTTGA
GAGAGAGGGTACCAAGGGAGAAGTACTCTCGAGAAGCTTT
CAGCAAAGTCCTGAGAACCTTCTT277NtFT3 ACTAGTGGATCCAAAGAATTC
ATGTTGAGAG AGAGGGTACCAGAAGTACTCTCGAGAAGCTT
TCACATATTG CATGGCTTAG277NtFT4 ACTAGTGGATCCAAAGAATTCAT
GCCGATACTGGGACAAGTAGAAGTACTCTCGAGAAGCTTTC
AGAACCTTCTTCCTCCGCA1.2.4 转基因植株的检测及表型观测
获得T2代转基因拟南芥植株后,各选取15株转基因植株进行表型观测,数据统计包括3个指标:各转基因植株从移栽到抽薹所需的时间(d)、抽薹时莲座叶的叶片数量和第一朵花开放的时间(d)。以转NtFT1和NtFT4拟南芥植株的莲座叶为试验材料,以野生型拟南芥为对照,利用荧光定量PCR的方法检测转NtFT1和NtFT4拟南芥植株中AtLFY、AtSOC1和AtAP1的表达情况。转基因拟南芥的各基因荧光定量引物见表4。
表 4 转基因拟南芥的不同基因qRT-PCR引物Table 4. Primers for qRT-PCR of different genes in transgenic A. thaliana引物名称
Primer name上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)Actin GCTGAGAGTTGATGGTGTGCT GGATACCCTTTCGCAGATAGAG AtLFY GTTAGGTTTTACGGCGAGCA GCAATCGTCTCCGTTCAGC AtAP1 ACCAAATCCAGCATCCTTACA TCAAGAGTCAGTTCGAGATCATTC AtSOC1 CTCTCAGTGCTTTGTGATGCT CGATTGAGCATGTTCCTATGCC 1.3 数据处理
采用GraphPad Prism软件进行数据处理及制图。
2. 结果与分析
2.1 中国水仙NtFT基因的克隆与生物信息学分析
对中国水仙转录组数据库进行筛选,鉴定得到4个NtFT家族成员。根据各基因的序列,设计特异引物,以中国水仙花芽cDNA为模板,利用RT-PCR技术克隆到4个FT基因 cDNA 的全长序列。经测序,NtFT1的开放阅读框(ORF)为525 bp,编码174个氨基酸;NtFT2的ORF为546 bp,编码181个氨基酸;NtFT3 的ORF为456 bp,编码151个氨基酸;NtFT4 的ORF为507 bp,编码168个氨基酸。
如图1所示,本研究中所克隆的FT基因编码的蛋白质都含有关键的氨基酸位点Y85。NtFT1、NtFT2和 NtFT3都与植物PEBP家族的成员一样,含有完整的DPDXP、GIHR、14个保守基序(Segment B)和LYN/IYN(图1)。NtFT3与其他FT基因的氨基酸序列同源性较差,特别是在C末端,没有LYN/IYN和GIHR保守基序,特别是FT基因的重要氨基酸Q140被S取代,这些改变可能会对它在中国水仙花发育中的功能产生一定影响。
系统发育分析结果表明,NtFT1属于FT-like IA类,与郁金香TgFT1和TgFT2、洋葱AcFT1和AcFT2聚类在一起;NtFT2、NtFT3和NtFT4与郁金香TgFT3蛋白和洋葱的3个FT同源蛋白AcFT3、AcFT5、AcFT6同属于FT-like II类(图2)。
2.2 NtFT基因在中国水仙不同组织部位中的表达分析
如图3所示,利用荧光定量PCR技术分析中国水仙FT基因在根、鳞茎盘、叶片、鳞片和花中的表达模式。结果表明,在不同器官中,NtFT基因的表达模式存在差异。从不同基因在各器官中的表达水平来看:在根中,NtFT4表达量最高,NtFT1最低;鳞茎盘和鳞片中各FT基因的表达模式相似,都是NtFT4的表达量最高,NtFT1最低;而在叶片中,NtFT2的表达量最高,NtFT4最低;在花中,NtFT3的表达量最高,NtFT4最低。从各基因在不同器官中的表达水平来看:NtFT1在花中表达量最高,在鳞茎盘中最低;NtFT2在叶片中表达量最高,在鳞茎盘中最低;NtFT3在花中表达量最高,在鳞茎盘中最低;NtFT4在鳞片中表达量最高,在叶片中最低。
2.3 NtFT基因在中国水仙花芽分化不同阶段的表达分析
分析4个FT基因在中国水仙花芽分化阶段的表达情况,结果(图4)发现NtFT1 在花芽分化过程中表达量呈先上升后下降的趋势,在7月30日表达量最高;NtFT2在主芽分化前期表达量的变化幅度不大,下降后又上升;NtFT3和NtFT4在整个花芽分化期间表达量都较低。
2.4 转NtFT基因拟南芥的开花表型观察
提取T0代抗生素阳性植株的DNA进行PCR检测,结果显示转NtFT基因抗生素阳性苗都有目的条带,而野生型拟南芥中没有检测到,说明各NtFT基因都分别转化到拟南芥中,获得转基因植株。从图5-A可以看出,过表达NtFT1和NtFT2拟南芥提早开花;过表达NtFT3拟南芥与野生型相比,在开花时间上没有明显差异;转NtFT4基因的拟南芥植株推迟开花。本试验统计了各转基因拟南芥和野生型拟南芥从移栽到抽薹时间、抽薹时莲座叶的数目和第一朵花开放的时间,发现在抽薹时,转NtFT1基因植株莲座叶数目比野生型少2.2片,抽薹和开花时间提前7.7 d和7.0 d,而且NtFT1转基因植株长势较弱;NtFT2过表达拟南芥植株花期稍微提前,抽薹和开花时间提前0.8 d和1.1 d;转NtFT3拟南芥在开花时间上与野生型没有明显差异,而NtFT4过表达拟南芥与野生型相比对拟南芥开花有抑制作用,转NtFT4基因植株莲座叶的数目比对照多1.8片,抽薹和开花时间会比对照晚3.5 d和3.3 d(图5)。
![]() 图 5 NtFT基因在拟南芥的异位表达A:过表达NtFT基因拟南芥开花情况;B:过表达 NtFT 基因拟南芥开花性状数据。*、**表示与WT相比差异显著(P<0.05)或极显著(P<0.01)。图6同。Figure 5. Ectopic expressions of NtFTs in A. thalianaA: Flowering of A. thaliana with overexpressed NtFT; B: Flowering traits data of A. thaliana with overexpressed NtFT; * and ** indicate significant difference(P<0.05) and (P<0.01), respectively. Same for Fig. 6.
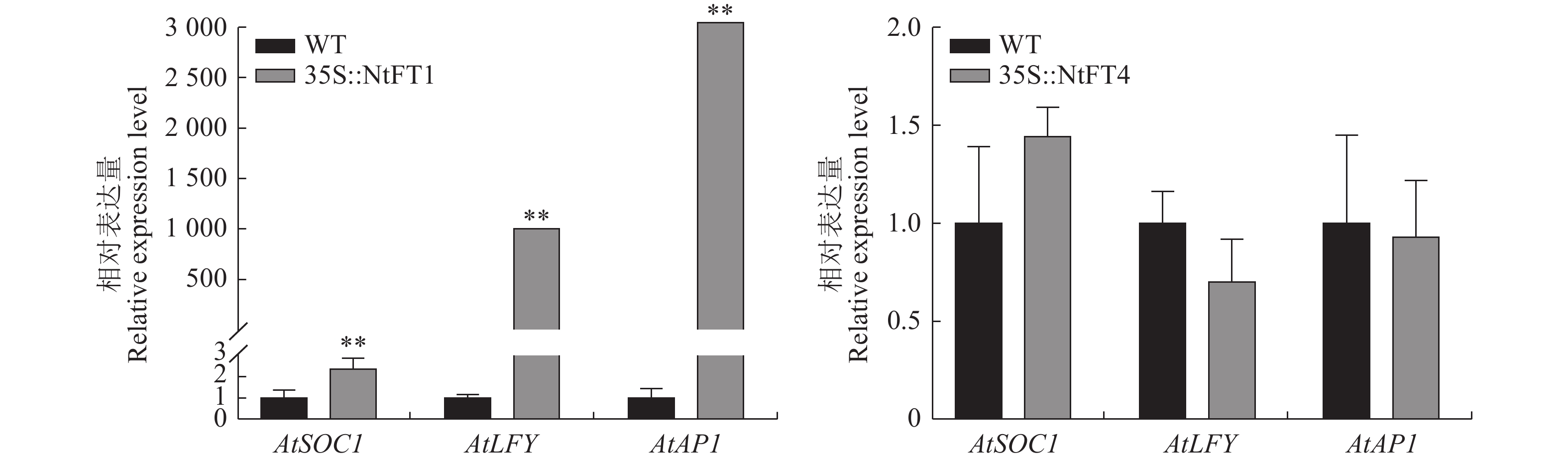
图 5 NtFT基因在拟南芥的异位表达A:过表达NtFT基因拟南芥开花情况;B:过表达 NtFT 基因拟南芥开花性状数据。*、**表示与WT相比差异显著(P<0.05)或极显著(P<0.01)。图6同。Figure 5. Ectopic expressions of NtFTs in A. thalianaA: Flowering of A. thaliana with overexpressed NtFT; B: Flowering traits data of A. thaliana with overexpressed NtFT; * and ** indicate significant difference(P<0.05) and (P<0.01), respectively. Same for Fig. 6.由于转NtFT1和NtFT4基因拟南芥表现出明显的提早或推迟开花,为了了解它们调控拟南芥开花的机理,利用实时荧光定量PCR技术检测了转基因植株AtLFY、AtSOC1和AtAP1的表达情况,结果显示,转NtFT1拟南芥的AtSOC1的表达量升高了2.7倍,而AtLFY和AtAP1基因表达量大幅上升,分别比野生拟南芥中各基因的表达量升高了1002和3040倍;转NtFT4拟南芥中AtSOC1的表达量是野生型的1.4倍,但AtLFY和AtAP1基因的表达量稍下降,分别为野生型的70.2%和92.9%(图6)。
3. 讨论
在植物的进化过程中,植物基因组复制加倍导致基因产生多拷贝现象。多拷贝的基因经常会发生功能分化或产生新功能,从而使植物更好地适应环境的变化[25]。在不同的植物中,FT基因的数目差异较大。如小鼠耳芥(Arabidopsis pumila)2个[26]、辣椒(Capsicum annuum) 4个[27]、月季(Rosa chinensis)2个[28]。单子叶植物的FT基因较多,Liu等[29]对17个单子叶植物和28个真双子叶植物基因组中FT基因进行分析,发现单子叶植物基因组中的FT亚家族平均14个,而真双子叶植物基因组中平均只有3个。由于水仙还未开展全基因组测序,本试验利用转录组测序结果通过RT-PCR技术克隆得到4个FT基因家族成员,表明中国水仙也与其他单子叶植物一样,具有多个FT基因。
对单子叶植物的FT基因的进化分析发现FT分为3个进化枝FT-like I、FT-like II和FT-like III,FT-like I又进化出于FT-like IA和FT-like IB亚支[29]。FT-like I可能是最保守的分支,代表了FT的祖先功能,调节单子叶植物的开花时间,还可能参与花器官的发育[29]。NtFT1属于FT-like IA类,与郁金香TgFT1[30]、洋葱AcFT1和AcFT2[31]聚类在一起,这些FT基因都具有促进植物开花的功能,表明NtFT1也可能具有促进植物开花的功能。NtFT1和NtFT2在水仙花芽分化过程中的花序原基(7月17日)和花原基(7月30日)表达量相对较高,花器官中也特异性表达,说明NtFT1可能是开花促进因子,异位转化拟南芥结果证明了这一点,转化NtFT1的拟南芥表现出提早开花。
FT-like II和FT-like III基因可能经历了功能多样化,如亚功能化、新功能化或功能丧失[29]。NtFT2、NtFT3、NtFT4与TgFT3[30]和AcFT3、AcFT5、AcFT6[31]同属于FT-like II类。这3个FT在中国水仙不同组织和花芽分化过程中表达模式并不一致,说明三者的功能可能也存在差异。NtFT2在中国水仙各组织器官中均有表达,而且在花芽分化过程中表达量较高但变化不大,这可能因为NtFT2虽然具有FT的保守基序,但调节开花的能力不强,因此转基因植株提早开花的表型不显著。这与百合(Lilium spp.)的LFT2相似,LFT2也在诱导期和分化期表达量都较高,转化拟南芥后也不能促进拟南芥提前开花。LFT2可能与鳞茎的发育有关[32],NtFT2是否也参与中国水仙的鳞茎发育,需要进一步验证。NtFT2和NtFT3亲缘关系较近,但在调控开花的功能上存在差异,可能是由于NtFT3的氨基酸组成发生的较大改变导致了这种差异。而且,这2个基因在表达模式上也存在差异,说明它们的功能可能发生了变化。NtFT4与郁金香的TgFT3亲缘关系较近,TgFT3对拟南芥的开花具有抑制作用[30],这与NtFT4转化拟南芥后的表型一致。
迄今为止,在多种植物中发现FT同源基因能够促进多种植物开花,如野菊花的CsFTL1、CsFT2和CsFTL3 [8−11]、苹果(Malus×domestica)的MdFT1和MdFT2[33]等,表明FT基因促进开花的功能保守。中国水仙NtFT1对拟南芥下游基因LFY、SOC1和AP1的表达影响分析,发现AtSOC1的表达量是野生型的2倍多,AtLFY和AtAP1的表达升高千倍,说明中国水仙NtFT1可能是通过促进AtSOC1、AtLFY和AtAP1的表达来促进拟南芥开花的,这与小叶杨(Populus simonii)FT基因作用机制相似[34]。一些植物的FT基因发生重复后产生的家族成员具有抑制植物开花的功能,如本试验的NtFT4、甜菜(Beta vulgaris)的BvFT2[35]、大豆(Glycine max)的GmFT2a和GmFT5a[36]。对过表达NtFT4植株的开花相关基因进行分析,发现拟南芥下游基因AtLFY和AtAP1的表达影响没有发生显著下降,而AtSOC1的表达量比野生型还要高,表明NtFT4抑制开花可能不是通过抑制这些基因的表达来实现的。
-
图 5 NtFT基因在拟南芥的异位表达
A:过表达NtFT基因拟南芥开花情况;B:过表达 NtFT 基因拟南芥开花性状数据。*、**表示与WT相比差异显著(P<0.05)或极显著(P<0.01)。图6同。
Figure 5. Ectopic expressions of NtFTs in A. thaliana
A: Flowering of A. thaliana with overexpressed NtFT; B: Flowering traits data of A. thaliana with overexpressed NtFT; * and ** indicate significant difference(P<0.05) and (P<0.01), respectively. Same for Fig. 6.
表 1 克隆中国水仙NtFT基因引物
Table 1 Primers for cloning NtFT of N. tazetta
引物名称
Primer name上游引物序列(5′-3′)
Upstream primer sequence (5′-3′)下游引物序列(5′-3′)
Downstream primer sequence (5′-3′)NtFT1 TTTCCGCTTATATCTCTTCTGGGAC TCGGGAAGTAGCAAGACGATCAAAC NtFT2 ATGTTGAGAGAGAGGGTACC TCAGCAAAGTCCTGAGAACCTTCTT NtFT3 GAAGTAGTCATGTTGAGAGAGAGG GTATCACATATTGCATGGCTTAGG NtFT4 GGTTAAGAGACAGAATGCCGAT TTTATGTCATTTATCGTCTG CTAG 表 2 FT基因表达qRT-PCR引物
Table 2 Primers for qRT-PCR of FT gene
引物名称
Primer name上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)上游引物序列(5’-3’)
Downstream primer sequence (5′-3′)NtActin GTTGACCCACCACTAAGAACAATG TGCCCAGAAGTGCTATTCCAG QNtFT1 CCAGCCAAAGGTTGAAGTCG CCCTGTGGTTCCTGGTATG QNtFT2 CTTATGAGAGCCCTCGAACACC CGCACACAGTTTGTTGAACTTCC QNtFT3 CGCACACAGTTTGTTGAACTTCC CACTGGTTGGTGACAGACATACC QNtFT4 TGGCAGGATGCGATGCAAGA CACGATGCGATGAATCCCCGA 表 3 pSAK277表达载体构建引物
Table 3 Primers for constructing pSAK277 expression vector
引物名称
Primer name上游引物序列(5′-3′)
Upstream primer sequence (5′-3′)下游引物序列(5′-3′)
Upstream primer sequence (5′-3′)277NtFT1 ACTAGTGGATCCAAAGAATTCATGA
GTAGGGATCCTTTGGTTATTGAGAAGTACTCTCGAGAAGCTT
TTAGGGGTACATCCTCCGGCCACCA277NtFT2 ACTAGTGGATCCAAAGAATTCAGTTGA
GAGAGAGGGTACCAAGGGAGAAGTACTCTCGAGAAGCTTT
CAGCAAAGTCCTGAGAACCTTCTT277NtFT3 ACTAGTGGATCCAAAGAATTC
ATGTTGAGAG AGAGGGTACCAGAAGTACTCTCGAGAAGCTT
TCACATATTG CATGGCTTAG277NtFT4 ACTAGTGGATCCAAAGAATTCAT
GCCGATACTGGGACAAGTAGAAGTACTCTCGAGAAGCTTTC
AGAACCTTCTTCCTCCGCA表 4 转基因拟南芥的不同基因qRT-PCR引物
Table 4 Primers for qRT-PCR of different genes in transgenic A. thaliana
引物名称
Primer name上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)上游引物序列(5’-3’)
Upstream primer sequence (5′-3′)Actin GCTGAGAGTTGATGGTGTGCT GGATACCCTTTCGCAGATAGAG AtLFY GTTAGGTTTTACGGCGAGCA GCAATCGTCTCCGTTCAGC AtAP1 ACCAAATCCAGCATCCTTACA TCAAGAGTCAGTTCGAGATCATTC AtSOC1 CTCTCAGTGCTTTGTGATGCT CGATTGAGCATGTTCCTATGCC -
[1] FORNARA F, DE MONTAIGU A, COUPLAND G. SnapShot: Control of flowering in Arabidopsis [J]. Cell, 2010, 141(3): 550−550.e2. DOI: 10.1016/j.cell.2010.04.024
[2] 张艺能, 周玉萍, 陈琼华, 等. 拟南芥开花时间调控的分子基础 [J]. 植物学报, 2014, 49(4):469−482. DOI: 10.3724/SP.J.1259.2014.00469 ZHANG Y N, ZHOU Y P, CHEN Q H, et al. Molecular basis of flowering time regulation in Arabidopsis [J]. Chinese Bulletin of Botany, 2014, 49(4): 469−482.(in Chinese) DOI: 10.3724/SP.J.1259.2014.00469
[3] KARLGREN A, GYLLENSTRAND N, KÄLLMAN T, et al. Evolution of the PEBP gene family in plants: Functional diversification in seed plant evolution [J]. Plant Physiology, 2011, 156(4): 1967−1977. DOI: 10.1104/pp.111.176206
[4] BANFIELD M J, BARKER J J, PERRY A C, et al. Function from structure? The crystal structure of human phosphatidylethanolamine-binding protein suggests a role in membrane signal transduction [J]. Structure, 1998, 6(10): 1245−1254. DOI: 10.1016/S0969-2126(98)00125-7
[5] VALVERDE F, MOURADOV A, SOPPE W, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering [J]. Science, 2004, 303(5660): 1003−1006. DOI: 10.1126/science.1091761
[6] 张乔松, 杨伟儿. 中国水仙花芽分化及贮藏期外界因子对花序数的影响 [J]. 园艺学报, 1987, 14(2):139−143,145. ZHANG Q S, YANG W E. On flower-bud differentiation of Chinese narcissus and the effect of external factors in storage on flower percentage [J]. Acta Horticulturae Sinica, 1987, 14(2): 139−143,145.(in Chinese)
[7] 申艳红, 姜涛, 赵湾湾, 等. 乙烯处理水仙催多花技术和机理的研究 [J]. 农业生物技术学报, 2019, 27(6):1003−1015. SHEN Y H, JIANG T, ZHAO W W, et al. Study on technology and mechanism of ethylene treatment promotes the formation of more flowers of Narcissus tazetta var. chinensis [J]. Journal of Agricultural Biotechnology, 2019, 27(6): 1003−1015.(in Chinese)
[8] ODA A, NARUMI T, LI T P, et al. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums [J]. Journal of Experimental Botany, 2012, 63(3): 1461−1477. DOI: 10.1093/jxb/err387
[9] MAO Y C, SUN J, CAO P P, et al. Functional analysis of alternative splicing of the FLOWERING LOCUS T orthologous gene in Chrysanthemum morifolium [J]. Horticulture Research, 2016, 3: 16058. DOI: 10.1038/hortres.2016.58
[10] WANG L J, SUN J, REN L P, et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum [J]. Plant Biotechnology Journal, 2020, 18(7): 1562−1572. DOI: 10.1111/pbi.13322
[11] SUN J, WANG H, REN L P, et al. CmFTL2 is involved in the photoperiod- and sucrose-mediated control of flowering time in chrysanthemum [J]. Horticulture Research, 2017, 4: 17001. DOI: 10.1038/hortres.2017.1
[12] OTAGAKI S, OGAWA Y, HIBRAND-SAINT OYANT L, et al. Genotype of FLOWERING LOCUS T homologue contributes to flowering time differences in wild and cultivated roses [J]. Plant Biology, 2015, 17(4): 808−815. DOI: 10.1111/plb.12299
[13] CHEN L, CAI Y P, QU M N, et al. Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T [J]. Plant, Cell & Environment, 2020, 43(4): 934−944.
[14] WU L, LI F, DENG Q H, et al. Identification and characterization of the FLOWERING LOCUS T/terminal flower 1 gene family in petunia [J]. DNA and Cell Biology, 2019, 38(9): 982−995. DOI: 10.1089/dna.2019.4720
[15] HELLER W P, YING Z T, DAVENPORT T L, et al. Identification of members of the Dimocarpus longan flowering locus T gene family with divergent functions in flowering [J]. Tropical Plant Biology, 2014, 7(1): 19−29. DOI: 10.1007/s12042-013-9134-0
[16] COELHO C P, MINOW M A A, CHALFUN-JÚNIOR A, et al. Putative sugarcane FT/TFL1 genes delay flowering time and alter reproductive architecture in Arabidopsis [J]. Frontiers in Plant Science, 2014, 5: 221.
[17] NAVARRO C, ABELENDA J A, CRUZ-ORÓ E, et al. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T [J]. Nature, 2011, 478(7367): 119−122. DOI: 10.1038/nature10431
[18] NIWA M, DAIMON Y, KUROTANI K I, et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis [J]. The Plant Cell, 2013, 25(4): 1228−1242. DOI: 10.1105/tpc.112.109090
[19] KINOSHITA T, ONO N, HAYASHI Y, et al. FLOWERING LOCUS T regulates stomatal opening [J]. Current Biology:CB, 2011, 21(14): 1232−1238. DOI: 10.1016/j.cub.2011.06.025
[20] CHEN M, PENFIELD S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time [J]. Science, 2018, 360(6392): 1014−1017. DOI: 10.1126/science.aar7361
[21] ANDRÉ D, MARCON A, LEE K C, et al. FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees [J]. Current Biology:CB, 2022, 32(13): 2988−2996.e4. DOI: 10.1016/j.cub.2022.05.023
[22] 陈洁. 水稻FT-Like基因OsFTL4的功能研究[D]. 扬州: 扬州大学, 2020. CHEN J. Function research of FT-like gene OsTTL4 in rice [D]. Yangzhou: Yangzhou University, 2020. (in Chinese)
[23] FENG Y, ZHU L Y, PAN T F, et al. Characterization of summer dormancy in Narcissus tazetta var. Chinensis and the role of NtFTs in summer dormancy and flower differentiation [J]. Scientia Horticulturae, 2015, 183: 109−117. DOI: 10.1016/j.scienta.2014.11.013
[24] LI X F, JIA L Y, XU J, et al. FT-like NFT1 gene may play a role in flower transition induced by heat accumulation in Narcissus tazetta var. chinensis [J]. Plant and Cell Physiology, 2013, 54(2): 270−281. DOI: 10.1093/pcp/pcs181
[25] CONANT G C, WOLFE K H. Turning a hobby into a job: How duplicated genes find new functions [J]. Nature Reviews Genetics, 2008, 9(12): 938−950. DOI: 10.1038/nrg2482
[26] 李永光, 金玉环, 郭力, 等. 小鼠耳芥PEBP基因家族全基因组鉴定及表达分析 [J]. 遗传, 2022, 44(1):80−94. LI Y G, JIN Y H, GUO L, et al. Genome-wide identification and expression analysis of the PEBP genes in Arabidopsis pumila [J]. Hereditas(Beijing), 2022, 44(1): 80−94.(in Chinese)
[27] 牛西强, 罗潇云, 康凯程, 等. 辣椒PEBP基因家族的全基因组鉴定、比较进化与组织表达分析 [J]. 园艺学报, 2021, 48(5):947−959. NIU X Q, LUO X Y, KANG K C, et al. Genome-wide identification, comparative evolution and expression analysis of PEBP gene family from Capsicum annuum [J]. Acta Horticulturae Sinica, 2021, 48(5): 947−959.(in Chinese)
[28] JIANG X D, ZHONG M C, DONG X, et al. Rosoideae-specific duplication and functional diversification of FT-like genes in Rosaceae [J]. Horticulture Research, 2022, 9: uhac059. DOI: 10.1093/hr/uhac059
[29] LIU H L, LIU X, CHANG X J, et al. Large-scale analyses of angiosperm Flowering Locus T genes reveal duplication and functional divergence in monocots [J]. Frontiers in Plant Science, 2023, 13: 1039500. DOI: 10.3389/fpls.2022.1039500
[30] LEEGGANGERS H A, ROSILIO-BRAMI T, BIGAS-NADAL J, et al. Tulipa gesneriana and Lilium longiflorum PEBP genes and their putative roles in flowering time control [J]. Plant and Cell Physiology, 2018, 59(1): 90−106. DOI: 10.1093/pcp/pcx164
[31] LEE R, BALDWIN S, KENEL F, et al. FLOWERING LOCUS T genes control onion bulb formation and flowering [J]. Nature Communications, 2013, 4: 2884. DOI: 10.1038/ncomms3884
[32] YAN X, CAO Q Z, HE H B, et al. Functional analysis and expression patterns of members of the FLOWERING LOCUS T (FT) gene family in Lilium [J]. Plant Physiology and Biochemistry, 2021, 163: 250−260. DOI: 10.1016/j.plaphy.2021.03.056
[33] KOTODA N, HAYASHI H, SUZUKI M, et al. Molecular characterization of FLOWERING LOCUS T-like genes of apple (Malus ×domestica borkh. ) [J]. Plant and Cell Physiology, 2010, 51(4): 561−575. DOI: 10.1093/pcp/pcq021
[34] 朱燕宇. 小叶杨FT基因家族的克隆及功能验证[D]. 南京: 南京林业大学, 2015. ZHU Y Y. Cloning and functional analysis of FT gene family from Populus simonii[D]. Nanjing: Nanjing Forestry University, 2015. (in Chinese)
[35] PIN P A, NILSSON O. The multifaceted roles of FLOWERING LOCUS T in plant development [J]. Plant, Cell & Environment, 2012, 35(10): 1742−1755.
[36] LIU W, JIANG B J, MA L M, et al. Functional diversification of Flowering Locus T homologs in soybean: GmFT1a and GmFT2a/5a have opposite roles in controlling flowering and maturation [J]. The New Phytologist, 2018, 217(3): 1335−1345. DOI: 10.1111/nph.14884
-
期刊类型引用(2)
1. 陈越,朱力琪,马春艳,戴宇. 富氢水对水仙花生长的影响. 中国农学通报. 2025(10): 89-95 .  百度学术
百度学术
2. 杨莉荟,王旭红,张馨然,王潇,彭少兵. 核桃开花基因JrAP1、JrFT的克隆与功能分析. 西北林学院学报. 2025(03): 55-64 .  百度学术
百度学术
其他类型引用(2)




 下载:
下载: