Bioinformatics of Growth-interacting Factor Genes in Foxtail Millet
-
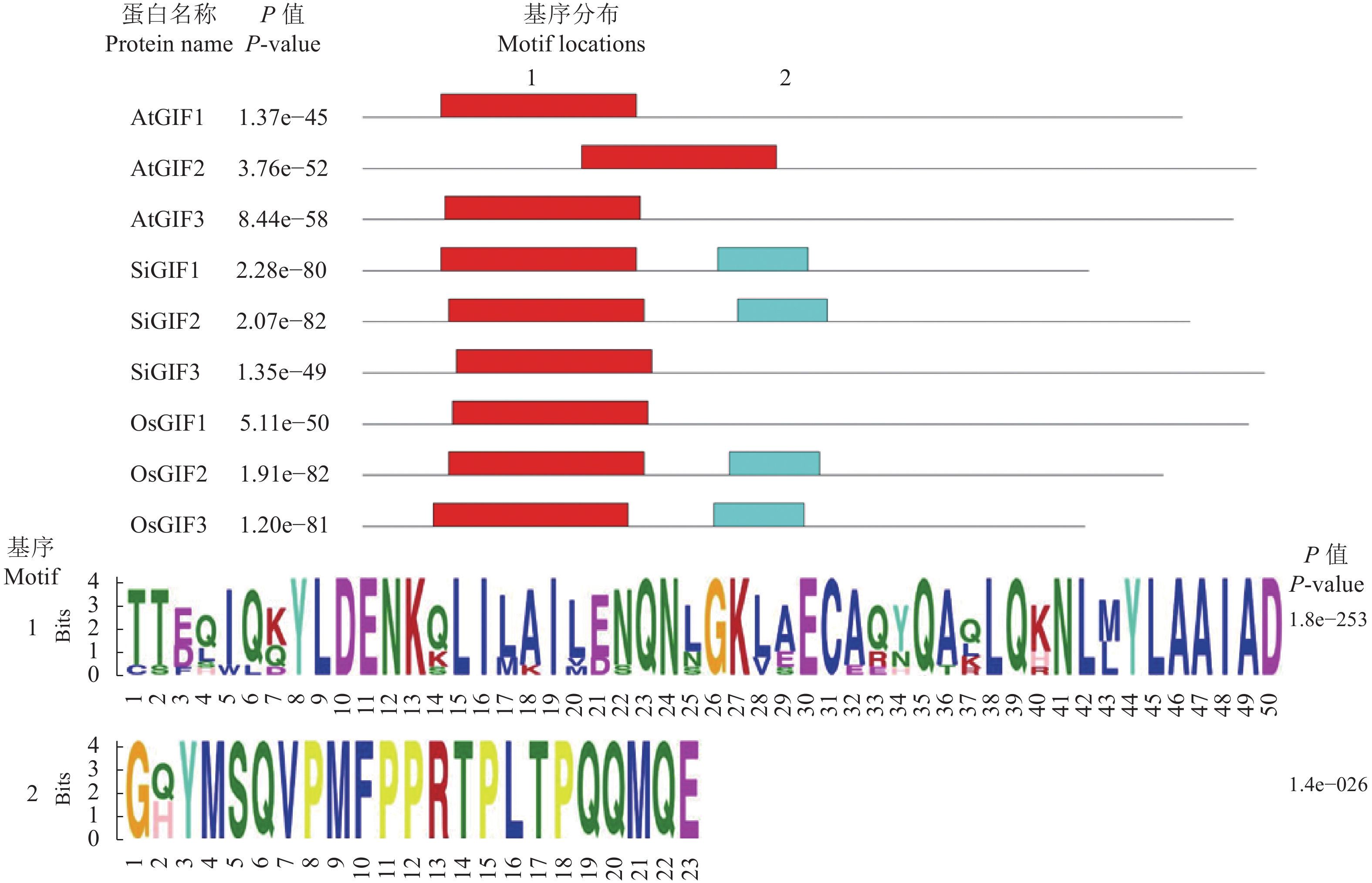
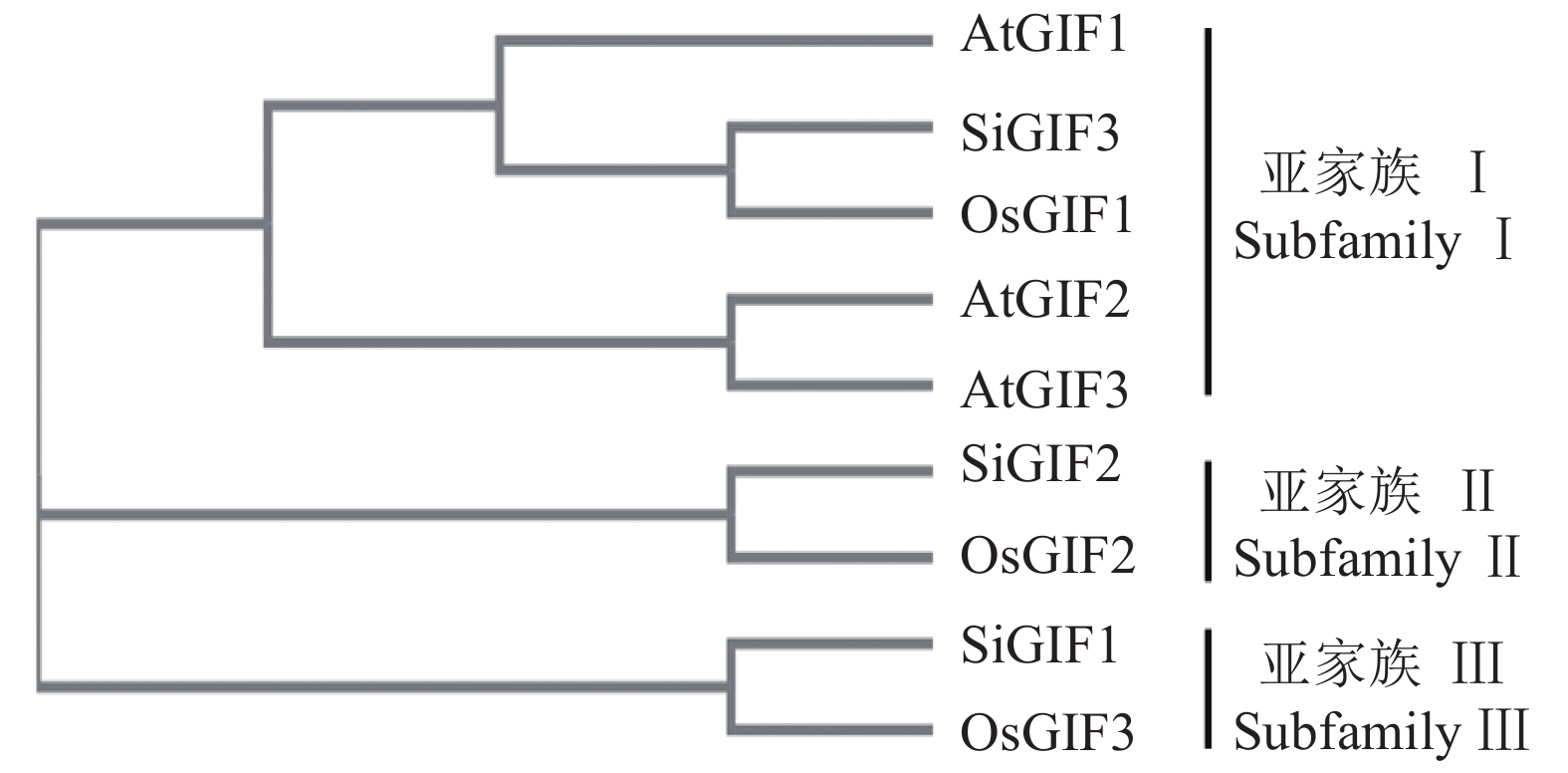
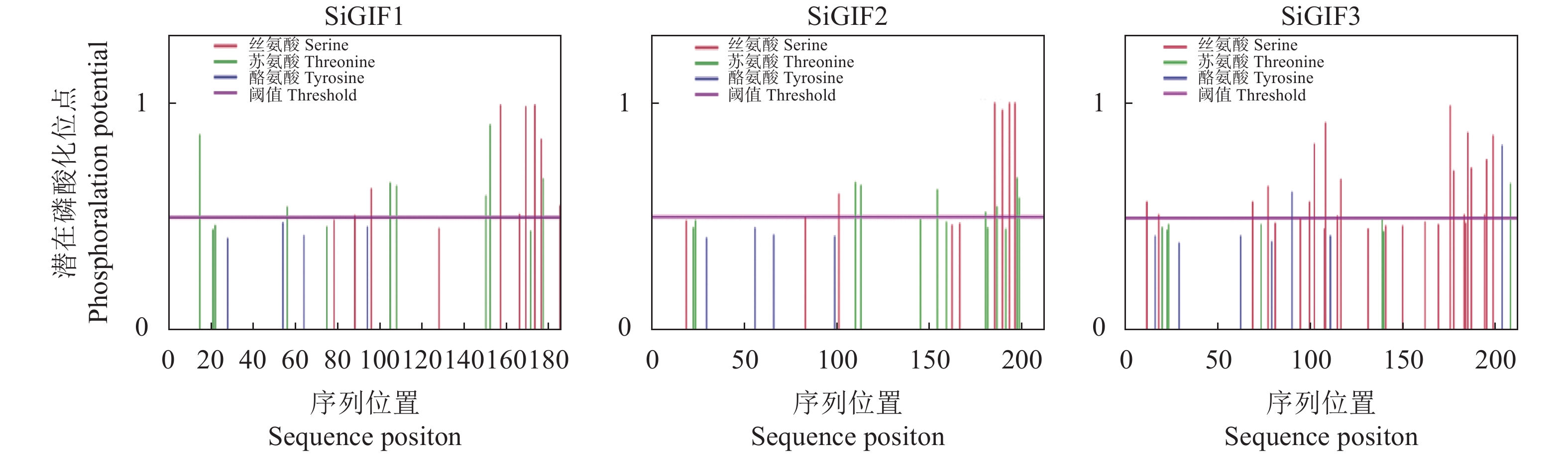
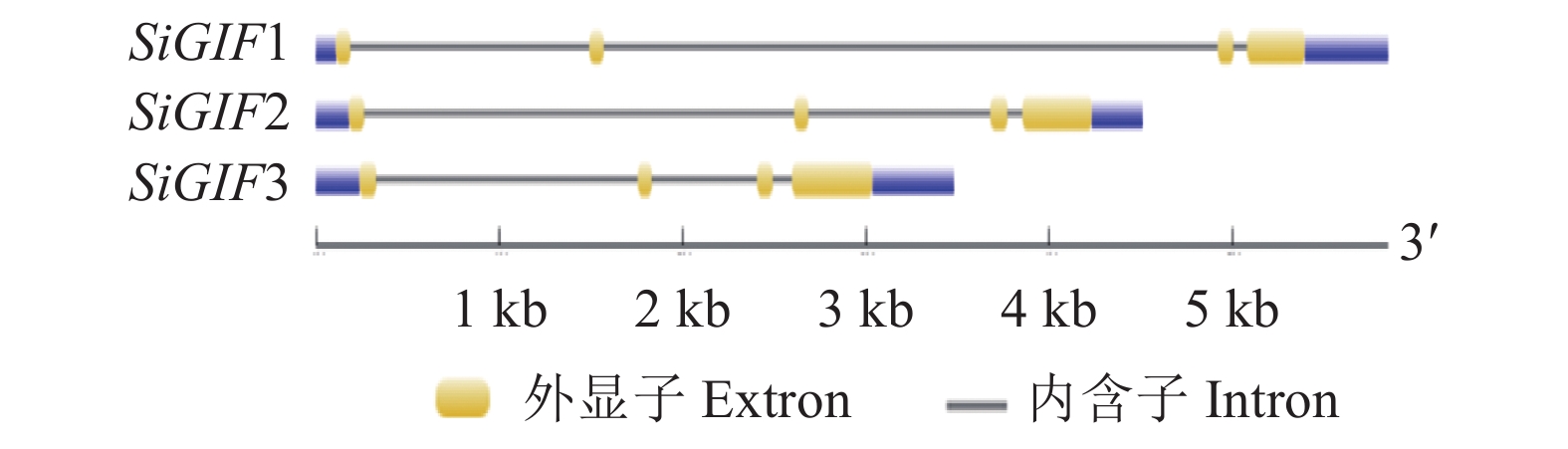
摘要:目的 生长调节因子互作因子(GRF-interacting factor,GIF)是植物体内一类转录共激活因子,在植物生长发育和逆境胁迫中起重要作用。通过系统分析谷子GIF基因家族的组成、各成员的结构以及进化关系,为GIF基因调节机制研究提供参考。方法 利用谷子基因组数据库,采用生物信息学的方法,鉴定谷子GIF 基因家族的基因结构、染色体定位,编码蛋白相似性、二级结构、跨膜区和磷酸化位点预测,通过序列比对进行进化和分类分析。结果 谷子含有3个GIF 基因,均含有4个外显子,分布于第3、8和9号染色体上。编码SiGIF1蛋白和SiGIF2蛋白相似性最高,为72.04%,SiGIF1蛋白和SiGIF3蛋白相似性最低,为37.08%。二级结构分析显示,谷子GIF蛋白无规则卷曲占比最高(41.56%~56.60%),其次为α-螺旋(34.43%~35.50%),再次为β-转角(5.19%~11.69%),β-折叠最低(3.23%~11.26%)。TMHMM 跨膜区进行分析显示,谷子GIF蛋白均不含有跨膜区。MEME保守基序分析显示,谷子GIF 蛋白均含有保守的SSXT (PF05030)结构域。磷酸化位点预测分析表明谷子GIF 蛋白均含有潜在磷酸化位点。结论 谷子GIF基因家族的基因结构、磷酸化位点预测等生物信息学分析结果将为揭示谷子GIF基因家族在谷子生长发育过程中的功能提供重要的线索。
-
关键词:
- 谷子 /
- 生长调节因子互作因子 /
- 基因家族 /
- 进化分析
Abstract:Objective The composition, structure, and evolution of each member of the growth-interacting factors (GIF) of the growth-regulating factors (GRF) and the transcription cofactors that closely associate with the growth, development, and stress response of plants in Setaria italica were analyzed.Method Based on the S. italica genome database and bioinformatics, the structure, characteristics, position on the chromosome, proteins similarity, secondary structure, transmembrane domain, and phosphorylation sites of the GIF genes were obtained.Result The 3 SiGIFs in S. italica genome contained 4 exons locating on the 3, 8, and 9 chromosomes. The greatest similarity between SiGIF1 and SiGIF2 was 72.04%, while the lowest was 37.08% between SiGIF1 and SiGIF3. The secondary structure consisted of 41.56%~56.60% random coils, 34.43%~35.50% alpha helix, 5.19%~11.69% beta turns and 3.23%~11.26% extended strands. The TMHMM transmembrane domain analysis showed no transmembrane domain in SiGIFs. MEME indicated that all SiGIFs contained conserved SSXT (PF05030) domain. And potential phosphorylation sites in the GIFs were predicted by analysis.Conclusion The bioinformatics revealed information on the structure, phosphorylation sites of SiGIF gene family provided crucial insights for the studies on the growth and development of plants.-
Keywords:
- Setaria italica /
- GIF /
- gene family /
- phylogeny analysis
-
-
表 1 谷子GIF基因家族的基本特征
Table 1 Characteristics of GIF genes in S. italica
基因名称
Gene name基因座
Locus name转录本
Transcript name别名
Alias染色体定位
Chromosome location开放阅读框
ORF/bp编码蛋白
Protein/aa外显子个数
Extron numberSiGIF1 Seita.3G360000 Seita.3G360000.1 Si023367m.g scaffold_3:46155062..46160910 + 561 186 4 SiGIF2 Seita.8G193300 Seita.8G193300.1 Si026850m.g scaffold_8:34062291..34066801− 639 212 4 SiGIF3 Seita.9G104500 Seita.9G104500.1 Si037326m.g scaffold_9:6299035..6302512− 696 231 4 表 2 谷子SiGIFs蛋白的二级结构分析
Table 2 Secondary structure of SiGIFs
蛋白名称
Protein nameα-螺旋
Alpha
helix/%β-折叠
Extended
strand/%β-转角
Beta
turn/%无规则卷曲
Random
coil/%SiGIF1 34.95 3.23 5.91 55.91 SiGIF2 34.43 3.77 5.19 56.60 SiGIF3 35.50 11.26 11.69 41.56 平均 Average 34.96 6.09 7.60 51.36 表 3 谷子SiGIFs蛋白潜在磷酸化位点分析
Table 3 Potential phosphorylation sites in
SiGIFs 谷子GIF 家族
SiGIF gene family位点数量 Site number 丝氨酸
Serine苏氨酸
Threoine酪氨酸
TyrosineSiGIF1 8 7 0 SiGIF2 5 7 0 SiGIF3 17 1 2 -
[1] DEBERNARDI J M, MECCHIA M A, VERCRUYSSEN L, et al. Post-transcriptional control of GRF transcription factors by microRNA miR396 and GIF co-activator affects leaf size and longevity [J]. Plant Journal, 2014, 79(3): 413−426. DOI: 10.1111/tpj.12567
[2] KIM J H. Biological roles and an evolutionary sketch of the GRF-GIF transcriptional complex in plants [J]. BMB Reports, 2019, 52(4): 227−238. DOI: 10.5483/BMBRep.2019.52.4.051
[3] LEE B H, KO J H, LEE S, et al. The Arabidopsis GRF-interacting factor gene family performs an overlapping function in determining organ size as well as multiple developmental properties [J]. Plant Physiology, 2009, 151(2): 655−668. DOI: 10.1104/pp.109.141838
[4] KIM J H, KENDE H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis [J]. PNAS, 2004, 101(36): 13374−13379. DOI: 10.1073/pnas.0405450101
[5] HORIGUCHI G, KIM G T, TSUKAYA H. The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana [J]. The Plant Journal, 2005, 43(1): 68−78. DOI: 10.1111/j.1365-313X.2005.02429.x
[6] VERCRUYSSEN L, VERKEST A, GONZALEZ N, et al. ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development [J]. The Plant Cell, 2014, 26(1): 210−229. DOI: 10.1105/tpc.113.115907
[7] LIANG G, HE H, LI Y, et al. Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis [J]. Plant Physiology, 2014, 164(1): 249−258. DOI: 10.1104/pp.113.225144
[8] ERCOLI M F, FERELA A, DEBERNARDI J M, et al. GIF transcriptional coregulators control root meristem homeostasis [J]. The Plant Cell, 2018, 30(2): 347−359. DOI: 10.1105/tpc.17.00856
[9] GAO F, WANG K, LIU Y, et al. Blocking miR396 increases rice yield by shaping inflorescence architecture [J]. Nature Plants, 2016, 2: 15196. DOI: 10.1038/nplants.2015.196
[10] NELISSEN H, EECKHOUT D, DEMUYNCK K, et al. Dynamic changes in ANGUSTIFOLIA3 complex composition reveal a growth regulatory mechanism in the maize leaf [J]. The Plant Cell, 2015, 27(6): 1605−1619. DOI: 10.1105/tpc.15.00269
[11] JIA G Q, HUANG X H, ZHI H, et al. A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica) [J]. Nature Genetics, 2013, 45(8): 957−961. DOI: 10.1038/ng.2673
[12] BENNETZEN J L, SCHMUTZ J, WANG H, et al. Reference genome sequence of the model plant Setaria [J]. Nature Biotechnology, 2012, 30(6): 555−561. DOI: 10.1038/nbt.2196
[13] ZHANG G Y, LIU X, QUAN Z W, et al. Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential [J]. Nature Biotechnology, 2012, 30(6): 549−554. DOI: 10.1038/nbt.2195
[14] YANG Z R, ZHANG H S, LI X K, et al. A mini foxtail millet with an Arabidopsis-like life cycle as a C4 model system [J]. Nature Plants, 2020, 6(9): 1167−1178. DOI: 10.1038/s41477-020-0747-7
[15] 张杰伟, 丁莉萍, 陈亚娟, 等. 杨树磷酸肌醇特异性磷脂酶C基因家族鉴定与分析 [J]. 福建农业学报, 2016, 31(11):1181−1186. ZHANG J W, DING L P, CHEN Y J, et al. Genome-wide analysis and identification of phosphoinositide-specific phospholipase C gene family in poplar(Populus trichocarpa) [J]. Fujian Journal of Agricultural Sciences, 2016, 31(11): 1181−1186.(in Chinese)
[16] BAILEY T L, JOHNSON J, GRANT C E, et al. The MEME suite [J]. Nucleic Acids Research, 2015, 43(W1): W39−W49. DOI: 10.1093/nar/gkv416
[17] GEOURJON C, DELÉAGE G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments [J]. Bioinformatics, 1995, 11(6): 681−684. DOI: 10.1093/bioinformatics/11.6.681
[18] SIEVERS F, WILM A, DINEEN D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega [J]. Molecular Systems Biology, 2011, 7: 539. DOI: 10.1038/msb.2011.75
[19] HUANG X Y, LIU G, ZHANG W W. Genome-wide analysis of LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Brassica rapa [J]. Brazilian Archives of Biology and Technology, 2018, 61: e18180049. DOI: 10.1590/1678-4324-2018180049
[20] ZHANG J B, WANG X P, WANG Y C, et al. Genome-wide identification and functional characterization of cotton (Gossypium hirsutum) MAPKKK gene family in response to drought stress [J]. BMC Plant Biology, 2020, 20(1): 1−14. DOI: 10.1186/s12870-019-2170-7
[21] MAO J X, ZHANG X S, ZHANG W J, et al. Genome-wide identification, characterization and expression analysis of the MITF gene in Yesso scallops (Patinopecten yessoensis) with different shell colors [J]. Gene, 2019, 688: 155−162. DOI: 10.1016/j.gene.2018.11.096
[22] LU Y Z, MENG Y L, ZENG J, et al. Coordination between GROWTH-REGULATING FACTOR1 and GRF-INTERACTING FACTOR1 plays a key role in regulating leaf growth in rice [J]. BMC Plant Biology, 2020, 20(1): 200. DOI: 10.1186/s12870-020-02417-0
[23] ZAN T, ZHANG L, XIE T T, et al. Genome-wide identification and analysis of the growth-regulating factor (GRF) gene family and GRF-interacting factor family in Triticum aestivum L [J]. Biochemical Genetics, 2020, 58(5): 705−724. DOI: 10.1007/s10528-020-09969-8
[24] ZHANG J W, ZHANG Z B, ZHU D, et al. Expression and initial characterization of a Phosphoinositide-specific phospholipase C from Populus tomentosa [J]. Journal of Plant Biochemistry and Biotechnology, 2015, 24(3): 338−346. DOI: 10.1007/s13562-014-0279-1
[25] LI S C, GAO F Y, XIE K L, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice [J]. Plant Biotechnology Journal, 2016, 14(11): 2134−2146. DOI: 10.1111/pbi.12569
[26] LIU W Y, ZHANG B, HE W Y, et al. Characterization of in vivo phosphorylation modification of differentially accumulated proteins in cotton fiber-initiation process [J]. Acta Biochimica et Biophysica Sinica, 2016, 48(8): 756−761. DOI: 10.1093/abbs/gmw055
[27] WANG Q, QIN G C, CAO M, et al. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants [J]. Nature Communications, 2020, 11(1): 679. DOI: 10.1038/s41467-020-14395-w
-
期刊类型引用(2)
1. 董晓静,夏启玉,降彦苗,刘国庆,程汝宏,赵辉. 两种PCR快速检测转基因糜子中外源基因的插入拷贝数. 热带农业科学. 2025(02): 55-63 .  百度学术
百度学术
2. 田春尧,冀慧玥,丁润月,郑乔木,周嘉裕,廖海. 转基因本氏烟草中CtDXR基因拷贝数及耐盐耐高温功能的初步分析. 生物学杂志. 2024(04): 17-23 .  百度学术
百度学术
其他类型引用(1)




 下载:
下载: