Bioinformatics analysis of differentially expressed genes in gosling plague and prediction of potential therapeutic Chinese medicine
-
摘要:目的
通过生物信息学方法筛选小鹅瘟(gosling plague, GP)相关数据集的致病核心基因及主要信号通路,预测潜在治疗靶点和有效干预中药。
方法通过收集GeneCards数据库中GP相关靶点并经Uniprot数据库标准化,并提取基因表达综合数据库(gene expression omnibus, GEO)肠道炎症数据集(GSE14841)和营养不良数据集(GSE43698),合并使用R语言Limmar包来筛选GP的差异表达基因(differentially expressed genes, DEGs)。利用DAVID数据库对DEGs进行基因本体论(Gene Ontology, GO)分析以及京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes, KEGG)富集分析,通过STRING数据库构建蛋白互作网络(protein-protein interaction network, PPI),Cytoscape 软件及其插件筛选子网络核心基因。将核心基因与Coremine Medical数据库相互映射,筛选能够治疗GP的潜在中药。
结果共筛选得到58个DEGs,富集分析结果显示,DEGs主要参与宿主细胞膜受体识别病毒蛋白、细胞质水解酶与转移酶活性等生物过程,定位于肌动蛋白细胞骨架(actin cytoskeleton, AC),环鸟苷酸-腺苷酸合成酶和干扰素基因刺激因子(cytosolic DNA-Sensing and the STING, cGAS-STING)、丝裂原活化蛋白激酶(mitogen-activated protein kinase, MARK)与Toll样受体(toll-like receptor, TLR)信号通路。通过PPI鉴定出Degree值前10名关键基因:干扰素诱导螺旋酶C结构域3(interferon induced with helicase C domain 3, IFIH3)、干扰素诱导螺旋酶C结构域1(interferon induced with helicase C domain 1, IFIH1)、线粒体抗病毒信号蛋白(mitochondrial antiviral signaling protein, MAVS)、C-C趋化因子配体5(C-C motif chemokine ligand 5, CCL5)、Toll样受体4(toll-like receptor 4, TLR4)、核因子κB(nuclear factor-kappa B, NF-κB)、Ras相关C3毒素底物2(ras-related C3 botulinum toxin substrate 2, RAC2)、Toll样受体9(toll-like receptor 9, TLR9)、早期生长应答基因1(early growth response 1, EGR1)、Erb-B2受体酪氨酸激酶3(erb-B2 receptor tyrosine kinase 3, ERBB3)。TLR4、TLR9、NF-κB、ERBB3为筛选到的GP感染性炎症4个核心基因。预测出治疗GP的潜在中药50种,中药类别主要包括清热解毒药、补虚药、解表药、收敛止泻药等4类。
结论本研究应用生物信息学方法明确了与GP相关的4个核心基因以及50种潜在靶向中药,为防治GP的天然药物研发提供了新思路与理论依据。
Abstract:ObjectiveBy using bioinformatics methods, we screened the core pathogenic genes and main signaling pathways of GP from related datasets, and predicted potential therapeutic targets and effective intervention Chinese herbal medicines.
MethodGP-related targets were collected from the GeneCards database and standardized via the Uniprot database. The intestinal inflammation dataset (GSE14841) and the malnutrition dataset (GSE43698) from the Gene Expression Omnibus (GEO) were extracted and combined. The differentially expressed genes (DEGs) of GP were screened using the Limma package in R language. The DAVID database was used for GO analysis and KEGG enrichment analysis of DEGs. The STRING database was used to construct the protein-protein interaction network (PPI), and the Cytoscape software and its plugins were used to screen the core genes of the sub-network. The core genes were mapped with the Coremine Medical database to screen for potential traditional Chinese medicines that could treat GP.
ResultA total of 58 DEGs were screened out. The results of enrichment analysis showed that the DEGs were mainly involved in biological processes such as the recognition of viral proteins by the host cell membrane receptors, and the activities of cytoplasmic hydrolases and transferases. They were located in the actin cytoskeleton (AC), the cytosolic DNA-Sensing and the STING (cGAS-STING), mitogen-activated protein kinase (MARK) and toll-like receptor (TLR) signaling pathways. Through the PPI, the top 10 key genes in terms of the Degree value were identified: interferon induced with helicase C domain 3 (IFIH3), interferon induced with helicase C domain 1 (IFIH1), mitochondrial antiviral signaling protein (MAVS), C-C motif chemokine ligand 5 (CCL5), toll-like receptor 4 (TLR4), nuclear factor-kappa B (NF-κB), ras-related C3 botulinum toxin substrate 2 (RAC2), toll-like receptor 9 (TLR9), early growth response 1 (EGR1), erb-B2 receptor tyrosine kinase 3 (ERBB3). TLR4, TLR9, NF-κB and ERBB3 were the four core genes identified in the inflammatory response to GP infection. A total of 50 potential traditional Chinese medicines for treating GP were predicted, and the categories of traditional Chinese medicines mainly included four types such as heat-clearing and detoxifying medicines, tonifying deficiency medicines, exterior-releasing medicines, and astringent antidiarrheal medicines.
ConclusionThis study applied bioinformatics methods to identify four core genes related to GP and 50 potential targeted traditional Chinese medicines, providing new ideas and theoretical basis for the development of natural medicines for the prevention and treatment of GP.
-
Keywords:
- gosling plague /
- goose parvovirus /
- potential Chinese herbs /
- virtual screening /
- bioinformatics
-
0. 引言
【研究意义】鹅细小病毒病又称小鹅瘟(gosling plague,GP)是由鹅细小病毒(goose parvovirus,GPV)引起的雏鹅和雏番鸭的一种高度接触性传染病,感染水禽出现不同程度腹泻、引起肠黏膜坏死脱落与纤维素性渗出性肠炎症为主要特征的疫病,发病率50%~70%、病死率40%~65%,病后耐过幼鹅及雏番鸭营养不良诱发发育迟缓[1]。GPV为细小病毒科细小病毒属成员,在宿主体内的主要定殖部位被认为是肠道上皮细胞,病毒粒子首先在肠道黏膜层大量复制并维持长时间存在,导致小肠发生纤维素性坏死性肠炎,从而紊乱肠道正常蠕动和肠管内物质代谢功能,引起患病水禽食欲减退、营养吸收障碍、免疫功能受损以及生长发育迟滞。另外,GPV还能通过肠壁血管扩散侵入血液循环到达宿主机体其他部位,在各组织器官中复制增殖并引起全身炎症反应,而且极易与其他肠道病毒混合感染,合并其他细菌继发侵袭,给有效免疫防控GP带来巨大困难与挑战[2−3]。因此,迫切需要开发新药物来减少GPV感染与传播。【前人研究进展】近年来,运用特定生物信息学工具与算法,依据设定的差异倍数和显著性水平等筛选条件,确定病毒感染宿主细胞中的差异表达基因,并从相关基因表达数据库获取病毒感染的疫病组和健康对照组数据,进行背景校正、标准化和数据转换等预处理,对筛选出的差异表达基因开展GO、KEGG等功能富集分析,探索基因潜在功能、参与的生物过程及相关信号通路,成为了鉴定各种病毒性疫病差异表达基因的有效方法[4]。研究表明,肠道炎症以及营养不良差异表达基因常与免疫炎症反应、细胞凋亡、信号转导等生物过程和通路紧密相关[5−6]。基于特定生物信息学平台或数据库,将筛选出的关键基因或差异表达基因与中药相关信息映射,分析中药中的活性成分及其与潜在靶点的相互作用,探讨中药治疗病毒感染的可能机制。中药凭借其多靶点、多组成、多通路、低毒副作用等特点展现出独特优势,特别是干预病毒复制增殖的作用近年来备受广泛关注,已成为新药物发现的宝贵资源[7]。大多数中药具有免疫双向调节功能,既可以增强机体的细胞免疫及体液免疫功能,也可以在某些情况下抑制免疫反应,减少炎性因子的释放[8],因此在抗GPV方面具有广阔的研究前景。【本研究切入点】目前鲜见有关基于生物信息学筛选小鹅瘟差异表达基因与预测潜在针对GPV感染治疗用中药的研究报道。【拟解决的关键问题】本研究以全面系统收集整理GP相关靶点数据为切入点,综合应用在线生物信息学研究方法分析和筛选GP相关的差异表达基因,进一步发掘核心基因,富集分析核心基因的功能和信号通路,靶向映射相关治疗用中药,从而探究GP的病理机制与治疗靶点,为干预GPV感染的天然药物开发与创制提供有力的理论支持。
1. 材料与方法
1.1 数据提取
在NCBI(https://www.ncbi.nlm.nih.gov/)、CTD(http://ctdbase.org/)和GeneCards(https:// www.genecards.org/)数据库中以goose、enteritis、diarrhea、growth retardation和gosling plague为关键词收集疫病相关靶点基因作为测试集,通过UniProt数据库对所得靶点进行标准化处理;从GEO[9]中获取肠道炎症(GSE14841)和营养不良(GSE43698)基因芯片数据作为验证集,其中GSE14841数据集包含4例健康和5例具有肠道炎症临床表达数据;GSE43698数据集包含4例健康和8例表现营养不良临床肌肉样本。利用GEO2R[10](https://www.ncbi.nlm.nih.gov/geo/geo2r/)进行在线分析,获取潜在的疾病靶点。最后,将上述数据库靶点进行去重、合并。为保证收集数据的可对比性,通过利用R-Studio 4.1.3软件对数据进行背景校正、标准化。
1.2 GP相关DEGs获取
引用R语言(http://www.r-project.org/)“limma”软件包[11],设定|log2 fold change|(|log2 FC|)≥0.5的基因为上调基因,|log2 FC|≤0.5的基因为下调基因和P<0.05为阈值条件,对下载平台文件与数据集后的样本基因进行差异分析和多重检验校正以获取DEGs,使用R语言软件ggplot2包[12]以及Complex Heatmap包[13]分别绘制差异表达基因的火山图和热图。对肠道炎症与营养不良合并数据集中筛选出的DEGs导入Venny 2.1(https://bioinfogp.cnb.csic.es/tools/venny/ index.html)取交集绘制韦恩图进行可视化。
1.3 GP相关DEGs功能注释和富集分析
将DEGs导入DAVID数据库[14](DAVID 6.8,http:// David.ncifcrf.gov)进行基因本体论(Gene Ontology, GO)功能、京都基因与基因组百科全书(Kyoto Encyclopedia of Genes and Genomes, KEGG)通路富集分析,获得每条功能/通路的基因列表及基因数目统计,应用超几何检验,得到显著富集的功能/通路以校正后的P<0.05为差异有统计学意义。GO功能富集分析包含生物过程(biological process, BP)、细胞组分(cell component, CC)和分子功能(molecular function, MF)相关的条目,KEGG是根据富集因子值分析核心靶点的作用通路,根据P<0.05和错误发现率(false discovery rate, FDR)<0.01筛选出重要的生物功能和作用通路,然后用Omicshare平台对结果进行可视化。
1.4 蛋白质-蛋白质相互作用网络构建及核心基因筛选
将共表达基因输入STRING数据库[15](https://string-db.org/)构建DEGs的蛋白质-蛋白质相互作用(protein-protein interaction network, PPI)网络,筛选标准为combined score≥0.4,预测靶标之间的相互作用关系,为深入认识GP的发病机制提供依据。将PPI网络的分析结果导入Cytoscape 3.9.1软件[16],应用Cytoscape的Network Analyzer功能对网络进行分析,根据Degree值大小调节节点的大小和颜色深浅对基因进行排序,Degree 值更高的基因更倾向于是关键基因。将Degree值较高的基因分别应用Cytoscape的cytoHubba[17]和Matthews correlation coefficient(MCC)算法[18](P<0.05,且表达趋势相同)以及MCODE插件[19](筛选条件为MCODE score>5,degree cutoff=2,node score cutoff=0.2,Haircut=true,Fluff=false,Max depth=100,K-score=2,P<0.05)进行模块分析,筛选得到核心基因。
1.5 靶点对接及中药预测
将筛选出的核心基因映射到Coremine Medical数据库[20](https://www.coremine.com/medical/)中,筛选出对GP具有潜在治疗作用的中药,以P<0.05作为统计学显著性结果依据。
1.6 统计学分析
所有统计学计算均采用R4.2.1及相关软件包完成。q值(q value)为娇正后的P值,表示P值产生假阳性的概率。相关性采用斯皮尔曼等级相关分析,P<0.05为差异具有统计学意义。
2. 结果与分析
2.1 差异表达基因的筛选
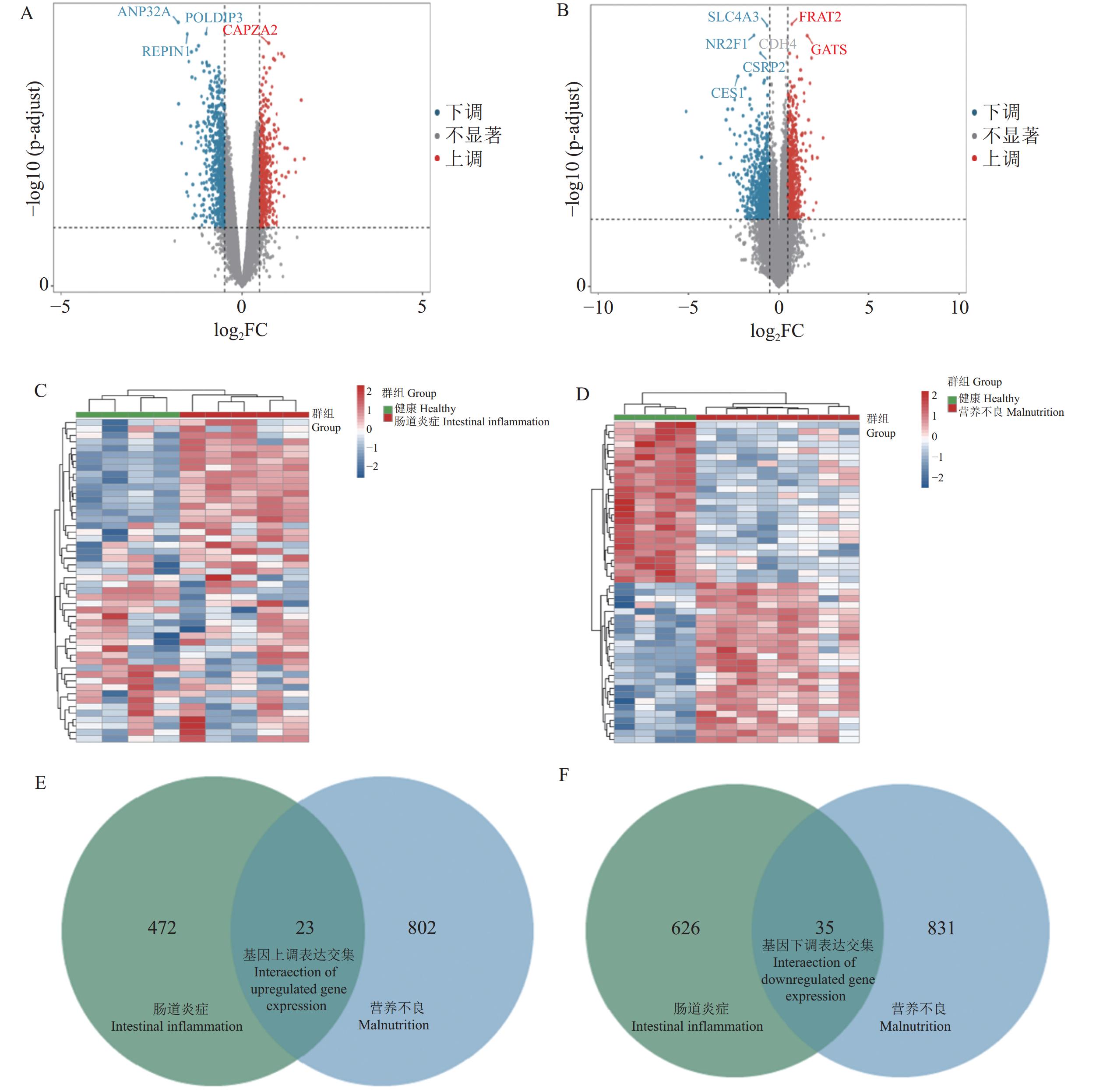
利用GEO2R对GP相关肠道炎症与营养不良数据集进行筛选,分别得到
5362 、4221 个符合条件的DEGs(图1A、B);分别按照|log2FC|大小排序获得两个数据集差异表达最显著的前25个上调、下调DEGs(图1C、D)。两个数据集共同DEGs有58个,其中包括上调基因23个、下调基因35个(图1E、F)。![]() 图 1 肠道炎症差异表达基因与营养不良差异表达基因筛选A~B:肠道炎症与营养不良相关差异基因表达火山图;C~D:肠道炎症与营养不良差异基因表达热图;E~F:肠道炎症与营养不良相关差异基因韦恩图。Figure 1. Intersection gene Venn diagram of differentially expressed genes in intestinal inflammation and malnutritionA–B: volcanic map of differentially expressed genes related to intestinal inflammation and malnutrition; C–D: heatmap of differentially expressed genes related to intestinal inflammation and malnutrition; E–F: Venn diagram of differentially expressed genes related to intestinal inflammation and malnutrition.
图 1 肠道炎症差异表达基因与营养不良差异表达基因筛选A~B:肠道炎症与营养不良相关差异基因表达火山图;C~D:肠道炎症与营养不良差异基因表达热图;E~F:肠道炎症与营养不良相关差异基因韦恩图。Figure 1. Intersection gene Venn diagram of differentially expressed genes in intestinal inflammation and malnutritionA–B: volcanic map of differentially expressed genes related to intestinal inflammation and malnutrition; C–D: heatmap of differentially expressed genes related to intestinal inflammation and malnutrition; E–F: Venn diagram of differentially expressed genes related to intestinal inflammation and malnutrition.2.2 共同差异基因表达的富集分析
GO富集分析显示,BP主要涉及肌动蛋白丝组织(actin filament organization)、蛋白质复合物组装的负调控(negative regulation of protein-containing complex assembly)、肌动蛋白细胞骨架组织的调控(regulation of actin cytoskeleton organization)、基于肌动蛋白丝过程的调控(regulation of actin filament-based process)、肌动蛋白丝帽(actin filament capping)、肌动蛋白丝解聚的负调控(negative regulation of actin filament depolymerization)、细胞因子产生的正向调控(positive regulation of cytokine production)等;CC主要涉及收缩纤维(contractile fiber)、细胞皮层(cell cortex)、线粒
体外膜(mitochondrial outer membrane)、细胞器外膜(organelle outer membrane)、外膜(outer membrane)、黏着斑(focal adhesion)、细胞-基质连接(cell-substrate junction)等;MF主要涉及肌动蛋白结合(actin binding)、钙调蛋白结合(calmodulin binding)、生长因子结合(growth factor binding)、磷脂酶激活活性(phospholipase activator activity)、脂肪酶激活活性(lipase activator activity)、蛋白酪氨酸激酶激活活性(protein tyrosine kinase activator activity)、唾液酸转移酶活性(sialyltransferase activity)(图2A)。KEGG分析结果表明,GP相关DEGs富集通路为:肌动蛋白细胞骨架调控(regulation of actin cytoskeleton)、血管平滑肌收缩(vascular smooth muscle contraction)、矿物质吸收(mineral absorption)、紧密连接(tight junction)、环鸟苷酸-腺苷酸合成酶和干扰素基因刺激因子(Cytosolic DNA-Sensing and the STING, cGAS-STING)信号通路、丝裂原活化蛋白激酶(mitogen-activated protein kinase, MARK)信号通路与Toll样受体(toll-like receptor, TLR)信号通路(图2B)。
2.3 筛选核心基因
在STRING数据库中导入DEGs构建PPI网络,使用Cytoscape软件的CytoHubba插件进行进一步分析,有40个基因彼此存在相互作用(图3A)。根据Degree值定义前10个关键基因为干扰素诱导螺旋酶C结构域3(interferon induced with helicase C domain 3, IFIH3)、干扰素诱导螺旋酶C结构域1(interferon induced with helicase C domain 1, IFIH1)、线粒体抗病毒信号蛋白(mitochondrial antiviral signaling protein, MAVS)、C-C趋化因子配体5(C-C motif chemokine ligand 5, CCL5)、Toll样受体4(toll-like receptor 4, TLR4)、核因子κB(nuclear factor-kappa B, NF-κB)、Ras相关C3毒素底物2(ras-related C3 botulinum toxin substrate 2, RAC2)、Toll样受体9(toll-like receptor 9, TLR9)、早期生长应答基因1(early growth response 1, EGR1)、Erb-B2受体酪氨酸激酶3(erb-B2 receptor tyrosine kinase 3, ERBB3)。进一步使用Cytoscape软件MCC算法以及MCODE插件对GP相关肠道炎症与营养不良数据集DEGs的上述10个关键基因进行模块分析取交集,结果表明:TLR4、TLR9、NF-κB、ERBB3在肠道炎症与营养不良验证集中均显示为上调基因。因此,确定TLR4、TLR9、NF-κB、ERBB3为GP感染性炎症的核心基因(图3B)。
2.4 中药预测结果
将4个核心基因映射到Coremine Medical数据库,以P<0.05为条件筛选干预GP的潜在治疗中药,共获得50种预测中药(表1),其中清热解毒药:黄连(Coptis chinensis Franch.)、茵陈(Artemisia capillaris Thunb.)、苦参(Sophora flavescens Ait.)、黄芩(Scutellaria baicalensis Georgi)、龙葵(Solanum nigrum L.)、土茯苓(Smilax glabra Roxb.)、黄柏(Phellodendron amurense Rupr.)、夏枯草(Prunella vulgaris L.)、天花粉(Trichosanthes kirilowii Maxim.)、茶树根(Camellia sinensis (L.) O. Ktze.)、枇杷叶(Eriobotrya japonica (Thunb.) Lindl.)、褐藻(Laminaria japonica Aresch.)、小叶海藻(Sargassum fusiforme (Harv.) Setch.)、矮地茶(Ardisia japonica (Thunb.) Blume)、枸骨叶(Ilex cornuta Lindl. et Paxt.)、墨旱莲(Eclipta prostrata (L.) L.)、桑白皮(Morus alba L.)、桑叶(Morus alba L.)、木蝴蝶(Oroxylum indicum (L.) Kurz)、没药(Commiphora myrrha Engl.),占40%;补虚药:山药(Dioscorea opposita Thunb.)、甘草(Glycyrrhiza uralensis Fisch.)、白术(Atractylodes macrocephala Koidz.)、人参(Panax ginseng C. A. Mey.)、党参(Codonopsis pilosula (Franch.) Nannf.)、大枣(Ziziphus jujuba Mill.)、薏苡仁(Coix lacryma-jobi L. var. ma-yuen (Roman.) Stapf)、白扁豆(Lablab purpureus (Linn.) Sweet)、黑芝麻(Sesamum indicum L.)、杜仲(Eucommia ulmoides Oliv.)、墨旱莲(Eclipta prostrata (L.) L.)、绞股蓝(Gynostemma pentaphyllum (Thunb.) Makino),占24%;解表药:葛根(Pueraria lobata(Willd.)Ohwi)、柴胡(Bupleurum chinense DC.)、鹅不食草(Centipeda minima (L.) A. Br. et Aschers)、葱白(Allium fistulosum L.),占8%;收敛止泻药:莲子肉(Nelumbo nucifera Gaertn. Fructus)、肉豆蔻(Myristica fragrans Houtt.)、诃子肉(Terminalia chebula Retz.),占6%。以上述4类中药为主,累计占比78%。
表 1 核心基因的中药预测结果Table 1. The prediction of traditional Chinese medicine corresponding to genes基因 Gene 中药 Traditional Chinese medicine TRL4 黄连、玫瑰花、山药、茵陈、苦参、甘草、莲子肉、白术、钩藤、夏枯草、
葛根、柴胡、鹅不食草、枳椇子、干姜、桑白皮、桑叶、杜仲TRL9 黄芩、甘草、人参、半夏、苦参 NF-κB 褐藻、木蝴蝶、木香、肉豆蔻、党参、柴胡、龙葵、薏苡仁、没药、吴茱萸、
甘草、土茯苓、钩藤、黄连、桑白皮、白果、桑叶、大枣、车前子、绞股蓝、
白扁豆、诃子肉、墨旱莲、杜仲、矮地茶、黄柏、枸骨叶ERBB3 小叶海藻、苦参、葱白、瓜蒌皮、夏枯草、麝香、天花粉、茶树根、枇杷叶、
瓜蒌、黑芝麻3. 讨论
GPV具有嗜肠性,主要在肠道黏膜上皮细胞内吸附、侵入并进行复制,导致肠道黏膜上皮细胞变性、坏死,微绒毛结构破坏,引发肠道炎症和损伤,严重影响肠道对营养物质的消化和吸收功能,患病雏鹅和雏番鸭生长发育受到严重阻碍[21]。GPV感染自然宿主后,可以被靶器官中细胞PRRs识别,引发一系列激酶的磷酸化级联反应,调节病毒感染细胞的增殖、分化和炎症反应,发挥细胞免疫和体液免疫功能来对抗病毒感染[22]。然而,GPV也可以利用炎症环境来促进自身的传播和复制,过度激活的炎症信号通路能够导致机体组织的病理损伤病变,延长GP病程[23]。目前对于GPV感染的分子机制的研究尚不明确,且GP确诊时大多数临床病例已是感染预后不良。因此,从基因层面探索GPV感染宿主后信号通路的生物标记物,挖掘中药药物,有助于GP的诊断和治疗。
本研究通过分析GP病例相关肠道炎症和营养不良数据集得到共同DEGs,总计58个,其中23个上调基因、35个下调基因。通过GO和KEGG富集分析探讨DEGs的生物学过程。GO富集分析显示,宿主天然免疫系统中胞质内生物和宿主衍生DNA感知cGAS-STING信号通路、细胞核内外生理信号传导MARK信号通路以及抗病毒免疫反应Toll样受体信号通路在GPV感染引起GP炎症病理和临床预后等方面均有关联[24]。KEGG富集分析表明,肌动蛋白作为细胞骨架上肌原纤维的重要组成部分,在细胞信号通路上传递机体生理病理级联信号中发挥重要作用,其在肠黏膜上皮细胞中的丰富程度,与肠道蠕动以及营养吸收功能密切相关,这种结构与功能的损伤或丧失都会导致炎症性肠炎[25−26]。
本研究根据Degree值选取前10个基因作为关键基因,分别为IFIH3、IFIH1、MAVS、CCL5、TLR4、NF-κB、RAC2、TLR9、EGR1、ERBB3。富集分析和PPI网络归纳提示,上调的DEGs(TLR4、TLR9、NF-κB、ERBB3)主要与病毒性肠炎以及营养吸收障碍相关,为GP感染性炎症的核心基因。TLRs属于模式识别受体(pattern recognition receptors, PRRs)家族,是参与非特异性免疫(天然免疫)的一类重要蛋白质分子,还是连接天然免疫和特异性免疫的桥梁,能够激活天然免疫细胞(如巨噬细胞、树突状细胞等),进而启动特异性免疫应答(如T细胞和B细胞介导的免疫应答)[27]。肠道炎症反应通常始于TLRs识别病原体相关分子模式(pathogen-associated molecular patterns, PAMPs)以及损伤相关分子模式(damage-associated molecular patterns, DAMPs),当肠道上皮细胞或固有层免疫细胞TLRs识别到病原微生物或其代谢产物时,会触发一系列信号级联反应,导致炎症因子的产生和释放[28]。TLRs的激活有助于清除病原体和修复受损组织;然而这种调控失效或炎症持续存在,则可能导致肠道屏障功能障碍和慢性炎症性疾病的发生[29]。因此,监测TLRs信号通路的活性变化,可以作为病毒性疫病的诊断和预后指标。在肠炎状态下,TLRs诱导的炎症反应依赖于MyD88接头分子介导的NF-κB信号转导途径[30]。NF-κB过度激活可能导致肠道上皮细胞异常增殖或凋亡抵抗,从而加剧肠道损伤[31]。目前抑制NF-κB信号通路已成为治疗肠炎的一种潜在策略,已有多种针对NF-κB信号通路的药物或治疗方法正在研发或临床试验中,如kappa B抑制因子激酶(inhibitor of kappa B kinase, IKK)抑制剂、抑制NF-κB活性蛋白(inhibitor of NF-κB, IκB)模拟物、NF-κB特异性抑制剂等。这些药物或治疗方法通过阻断NF-κB信号通路的激活、减少促炎细胞因子的产生,从而减轻肠道炎症反应[32]。ERBB3属于表皮生长因子受体(epidermal growth factor receptor, EGFR/ERBB)家族,是一种重要的细胞膜受体酪氨酸激酶,作为信号转导通路中的关键分子,在肠上皮、骨骼、肌肉和皮肤中均有表达,可以正反馈肠道炎症的严重程度,其抑制剂在动物模型中显示出对肠道炎症的缓解作用[33]。
GP病程复杂且症状多样,仅有少数患病水禽可以自愈,但生长发育受到严重阻碍。在国家“减抗替抗”的政策背景下,绿色保健药物抗GPV感染已经被众多养殖户所接受。中药具有多成分、多靶点、多途径、作用温和、不良反应小,在帮助水禽抵抗病毒和其他病原体侵染中得到了广泛认可[34]。由板蓝根、金银花、黄芩、柴胡、官桂、赤石脂、生地、赤芍、水牛角组成的中药方剂,可增强肠道免疫,其主要机制是包括调节炎症因子、保护肠道黏膜和改善肠道菌群[35];当归、白芨、大青叶、板蓝根、紫花地丁、绿豆和甘草等中药组成复方中药口服液可以能够有效提高感染GPV雏鹅的免疫和抗氧化功能,封闭宿主细胞表面特异性结合受体,阻滞病毒吸附或抑制病毒在细胞内复制增殖[36−38]。另外,部分海洋天然药物也可以通过激发调动宿主免疫防御系统来间接发挥抗肠道病毒感染的作用,促进受损肠道黏膜愈合,增强肠道物理屏障,降低炎症因子表达,调理并改善病毒性免疫病理反应引起的肠道菌群失衡与营养吸收障碍[39−40]。
本研究基于虚拟筛选以及相关生物信息学技术,预测出具有抑制GPV感染并治疗GP引起的肠道炎症和营养不良的潜在中药,其主要作用机制可能为多种活性成分通过TLR4、TLR9、NF-κB与ERBB3等基因靶点,抑制炎症反应、修复免疫失衡、调节肠道菌群组成、抗病毒等途径,缓解GPV感染所致GP疫情,从而维持水禽个体健康所需的代谢和免疫平衡。综上所述,通过挖掘GP发生相关的核心基因,有利于探索GPV感染的发病机制,为相关药物设计提供新思路。
-
图 1 肠道炎症差异表达基因与营养不良差异表达基因筛选
A~B:肠道炎症与营养不良相关差异基因表达火山图;C~D:肠道炎症与营养不良差异基因表达热图;E~F:肠道炎症与营养不良相关差异基因韦恩图。
Figure 1. Intersection gene Venn diagram of differentially expressed genes in intestinal inflammation and malnutrition
A–B: volcanic map of differentially expressed genes related to intestinal inflammation and malnutrition; C–D: heatmap of differentially expressed genes related to intestinal inflammation and malnutrition; E–F: Venn diagram of differentially expressed genes related to intestinal inflammation and malnutrition.
表 1 核心基因的中药预测结果
Table 1 The prediction of traditional Chinese medicine corresponding to genes
基因 Gene 中药 Traditional Chinese medicine TRL4 黄连、玫瑰花、山药、茵陈、苦参、甘草、莲子肉、白术、钩藤、夏枯草、
葛根、柴胡、鹅不食草、枳椇子、干姜、桑白皮、桑叶、杜仲TRL9 黄芩、甘草、人参、半夏、苦参 NF-κB 褐藻、木蝴蝶、木香、肉豆蔻、党参、柴胡、龙葵、薏苡仁、没药、吴茱萸、
甘草、土茯苓、钩藤、黄连、桑白皮、白果、桑叶、大枣、车前子、绞股蓝、
白扁豆、诃子肉、墨旱莲、杜仲、矮地茶、黄柏、枸骨叶ERBB3 小叶海藻、苦参、葱白、瓜蒌皮、夏枯草、麝香、天花粉、茶树根、枇杷叶、
瓜蒌、黑芝麻 -
[1] HUO X R,CHEN Y M,ZHU J R,et al. Evolution,genetic recombination,and phylogeography of goose parvovirus[J]. Comparative Immunology,Microbiology and Infectious Diseases,2023,102:102079.
[2] WANG H Z,LEI D,XU B Y,et al. Continuous surveillance of pathogens detects excretion of avian orthoreovirus and parvovirus by several wild waterfowl:Possible wild bird reservoirs[J]. Poultry Science,2024,103(8) :103940.
[3] ZHANG S Z,DONG H,LIN F Q,et al. Development and application of a multiplex PCR method for the simultaneous detection of goose parvovirus,waterfowl reovirus,and goose astrovirus in Muscovy ducks[J]. Journal of Virological Methods,2024,324:114857. DOI: 10.1016/j.jviromet.2023.114857
[4] ONISIFOROU A,SPYROU G M. Identification of viral-mediated pathogenic mechanisms in neurodegenerative diseases using network-based approaches[J]. Briefings in Bioinformatics,2021,22(6) :bbab141. DOI: 10.1093/bib/bbab141
[5] STRYSKO J P,STEENHOFF A P. Biomarkers make the case for a comprehensive approach to diagnosing severe acute malnutrition[J]. Pediatrics,2021,147(6) :e2021050215.
[6] HU C Q,GE M,LIU Y,et al. From inflammation to depression:Key biomarkers for IBD-related major depressive disorder[J]. Journal of Translational Medicine,2024,22(1) :997.
[7] WANG B,WANG X F,XIONG Z W,et al. A review on the applications of Traditional Chinese medicine polysaccharides in drug delivery systems[J]. Chinese Medicine,2022,17(1) :12.
[8] SINGH S,MURTI Y,SEMWAL B. Antiviral activity of natural herbs and their isolated BioactiveCompounds:A review[J]. Combinatorial Chemistry & High Throughput Screening,2024,27(14) :2013−2042.
[9] CLOUGH E,BARRETT T,WILHITE S E,et al. NCBI GEO Archive for gene expression and epigenomics data sets:23-year update[J]. Nucleic Acids Research,2024,52(D1) :D138−D144.
[10] LIU X X,ZHANG X H,LI L Q,et al. Bioinformatics analysis of potential key genes and pathways in neonatal necrotizing enterocolitis[J]. BMC Pediatrics,2022,22(1) :658.
[11] RITCHIE M E,PHIPSON B,WU D,et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies[J]. Nucleic Acids Research,2015,43(7) :e47. DOI: 10.1093/nar/gkv007
[12] GUSTAVSSON E K,ZHANG D,REYNOLDS R H,et al. Ggtranscript:An R package for the visualization and interpretation of transcript isoforms using ggplot2[J]. Bioinformatics,2022,38(15) :3844−3846. DOI: 10.1093/bioinformatics/btac409
[13] GU Z G,HÜBSCHMANN D. Make interactive complex heatmaps in R[J]. Bioinformatics,2022,38(5) :1460−1462. DOI: 10.1093/bioinformatics/btab806
[14] HUANG D W,SHERMAN B T,LEMPICKI R A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources[J]. Nature Protocols,2009,4(1) :44−57. DOI: 10.1038/nprot.2008.211
[15] SZKLARCZYK D,KIRSCH R,KOUTROULI M,et al. The STRING database in 2023:Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest[J]. Nucleic Acids Research,2023,51(D1) :D638−D646. DOI: 10.1093/nar/gkac1000
[16] SHANNON P,MARKIEL A,OZIER O,et al. Cytoscape:A software environment for integrated models of biomolecular interaction networks[J]. Genome Research,2003,13(11) :2498−2504. DOI: 10.1101/gr.1239303
[17] CHIN C H,CHEN S H,WU H H,et al. cytoHubba:Identifying hub objects and sub-networks from complex interactome[J]. BMC Systems Biology,2014,8(S4) :S11. DOI: 10.1186/1752-0509-8-S4-S11
[18] CHICCO D,JURMAN G. A statistical comparison between Matthews correlation coefficient (MCC) ,prevalence threshold,and Fowlkes–Mallows index[J]. Journal of Biomedical Informatics,2023,144:104426. DOI: 10.1016/j.jbi.2023.104426
[19] BADER G D,HOGUE C W V. An automated method for finding molecular complexes in large protein interaction networks[J]. BMC Bioinformatics,2003,4:2. DOI: 10.1186/1471-2105-4-2
[20] 吴娜,饶颖,周佳林,等. 中药通过调节铁死亡作用治疗溃疡性结肠炎的用药规律研究[J]. 中成药,2023,45(10) :3482−3486. DOI: 10.3969/j.issn.1001-1528.2023.10.056 WU N,RAO Y,ZHOU J L,et al. Study on the law of traditional Chinese medicine in treating ulcerative colitis by regulating iron death[J]. Chinese Traditional Patent Medicine,2023,45(10) :3482−3486. (in Chinese) DOI: 10.3969/j.issn.1001-1528.2023.10.056
[21] NIU Y J,ZHAO L L,LIU B H,et al. Comparative genetic analysis and pathological characteristics of goose parvovirus isolated in Heilongjiang,China[J]. Virology Journal,2018,15(1) :27. DOI: 10.1186/s12985-018-0935-5
[22] YANG Y T,DENG Z C,ZHANG L J,et al. Novel goose parvovirus VP1 targets IRF7 protein to block the type I interferon upstream signaling pathway[J]. Poultry Science,2024,103(9) :104065. DOI: 10.1016/j.psj.2024.104065
[23] LIU M D,XIANG F J,PAN J L,et al. Host-derived lactic acid bacteria alleviate short beak and dwarf syndrome by preventing bone loss,intestinal barrier disruption,and inflammation[J]. Veterinary Microbiology,2024,296:110187. DOI: 10.1016/j.vetmic.2024.110187
[24] LIN C,ZHENG M,XIAO S F,et al. Duck cGAS inhibits DNA and RNA virus replication by activating IFNs and antiviral ISGs[J]. Frontiers in Immunology,2023,14:1101335. DOI: 10.3389/fimmu.2023.1101335
[25] HU S,SEVIER C S,KURPIOS N A. Protocol to detect smooth muscle actin-alpha and measure oxidative damage in neonatal mouse intestine[J]. STAR Protocols,2022,3(3) :101524. DOI: 10.1016/j.xpro.2022.101524
[26] MORI T,GIOVANNELLI L,BILIA A R,et al. Exosomes:Potential next-generation nanocarriers for the therapy of inflammatory diseases[J]. Pharmaceutics,2023,15(9) :2276. DOI: 10.3390/pharmaceutics15092276
[27] FAGHFOURI A H,ZARRIN R,MALEKI V,et al. A comprehensive mechanistic review insight into the effects of micronutrients on toll-like receptors functions[J]. Pharmacological Research,2020,152:104619. DOI: 10.1016/j.phrs.2019.104619
[28] ARANCIBIA S A,BELTRÁN C J,AGUIRRE I M,et al. Toll-like receptors are key participants in innate immune responses[J]. Biological Research,2007,40(2) :97−112.
[29] KUZMICH N N,SIVAK K V,CHUBAREV V N,et al. TLR4 signaling pathway modulators as potential therapeutics in inflammation and sepsis[J]. Vaccines,2017,5(4) :34. DOI: 10.3390/vaccines5040034
[30] BARNABEI L,LAPLANTINE E,MBONGO W,et al. NF-κB:At the borders of autoimmunity and inflammation[J]. Frontiers in Immunology,2021,12:716469. DOI: 10.3389/fimmu.2021.716469
[31] NTUNZWENIMANA J C,BOUCHER G,PAQUETTE J,et al. Functional screen of inflammatory bowel disease genes reveals key epithelial functions[J]. Genome Medicine,2021,13(1) :181. DOI: 10.1186/s13073-021-00996-7
[32] YU H,LIN L B,ZHANG Z Q,et al. Targeting NF-κB pathway for the therapy of diseases:Mechanism and clinical study[J]. Signal Transduction and Targeted Therapy,2020,5(1) :209. DOI: 10.1038/s41392-020-00312-6
[33] HICKS M R,HISERODT J,PARAS K,et al. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs[J]. Nature Cell Biology,2018,20(1) :46−57. DOI: 10.1038/s41556-017-0010-2
[34] LI L,YANG L L,YANG L Q,et al. Network pharmacology:A bright guiding light on the way to explore the personalized precise medication of traditional Chinese medicine[J]. Chinese Medicine,2023,18(1) :146. DOI: 10.1186/s13020-023-00853-2
[35] 黄爱英. 小鹅瘟的防治[J]. 畜禽业,2023,34(7) :83−85. HUANG A Y. Prevention and control of gosling plague[J]. Livestock and Poultry Industry,2023,34(7) :83−85. (in Chinese)
[36] 于浩然,李天琦,王连英,等. 复方中草药口服液对感染小鹅瘟病毒雏鹅免疫和抗氧化功能的影响[J]. 黑龙江畜牧兽医,2018(16) :162−164 YU H R,LI T Q,WANG L Y,et al. The effect of oral liquid of compound Chinese herbal medicine on immune and antioxidant functions of goslings infected with Goose parvovirus[J]. Heilongjiang Animal Science and Veterinary Medicine,2018(16) :162−164. (in Chinese)
[37] CHEN Z,YE S Y. Research progress on antiviral constituents in traditional Chinese medicines and their mechanisms of action[J]. Pharmaceutical Biology,2022,60(1) :1063−1076. DOI: 10.1080/13880209.2022.2074053
[38] 李天琦. 香菇多糖和黄芪多糖对感染GPV雏鹅免疫及小肠炎性损伤的作用[D]. 哈尔滨:东北农业大学,2019. LI T Q. Effects of Astragalus and Lentinus polysaccharides on immunity and small intestinal injury of gosling infected by GPV. Harbin:Northeast Agricultural University,2019. (in Chinese)
[39] AKBARI A,BIGHAM A,RAHIMKHOEI V,et al. Antiviral polymers:A review[J]. Polymers,2022,14(9) :1634. DOI: 10.3390/polym14091634
[40] YANG Y,XIAO G,CHENG P,et al. Protective application of Chinese herbal compounds and formulae in intestinal inflammation in humans and animals[J]. Molecules,2023,28(19) :6811. DOI: 10.3390/molecules28196811




 下载:
下载:


