Cloning and Expressions of CmCCR1 in Citrus maxima Sanhongmiyou
-
摘要:目的
肉桂酰辅酶A还原酶(cinnamoyl-CoA reductase,CCR)基因是木质素合成的关键基因,本研究旨在探明该基因在柚汁胞粒化过程中的作用机制。
方法通过生物信息学、基因克隆与亚细胞定位、荧光定量PCR技术,筛选克隆三红蜜柚CmCCR基因,及其在两个品种(三红蜜柚和六月早1号柚)不同发育时期果实汁胞中的表达情况,并利用乙酰溴法测定柚汁胞木质素含量。
结果从三红蜜柚汁胞转录组数据中筛选得到5个CmCCR基因,根据果实汁胞成熟期木质素含量与CmCCRs基因的转录水平变化的相关性,初步明确CmCCR1可能参与汁胞木质素代谢;克隆所得CmCCR1基因序列,其开放阅读框长度为
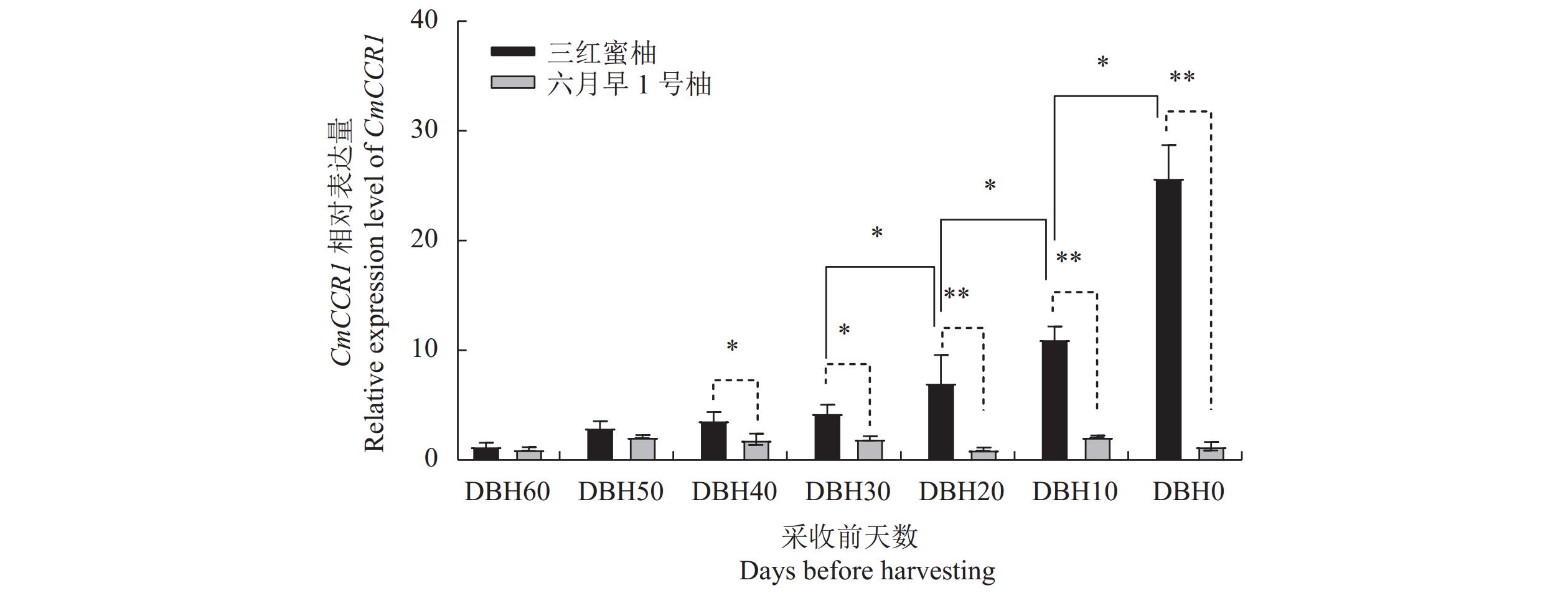
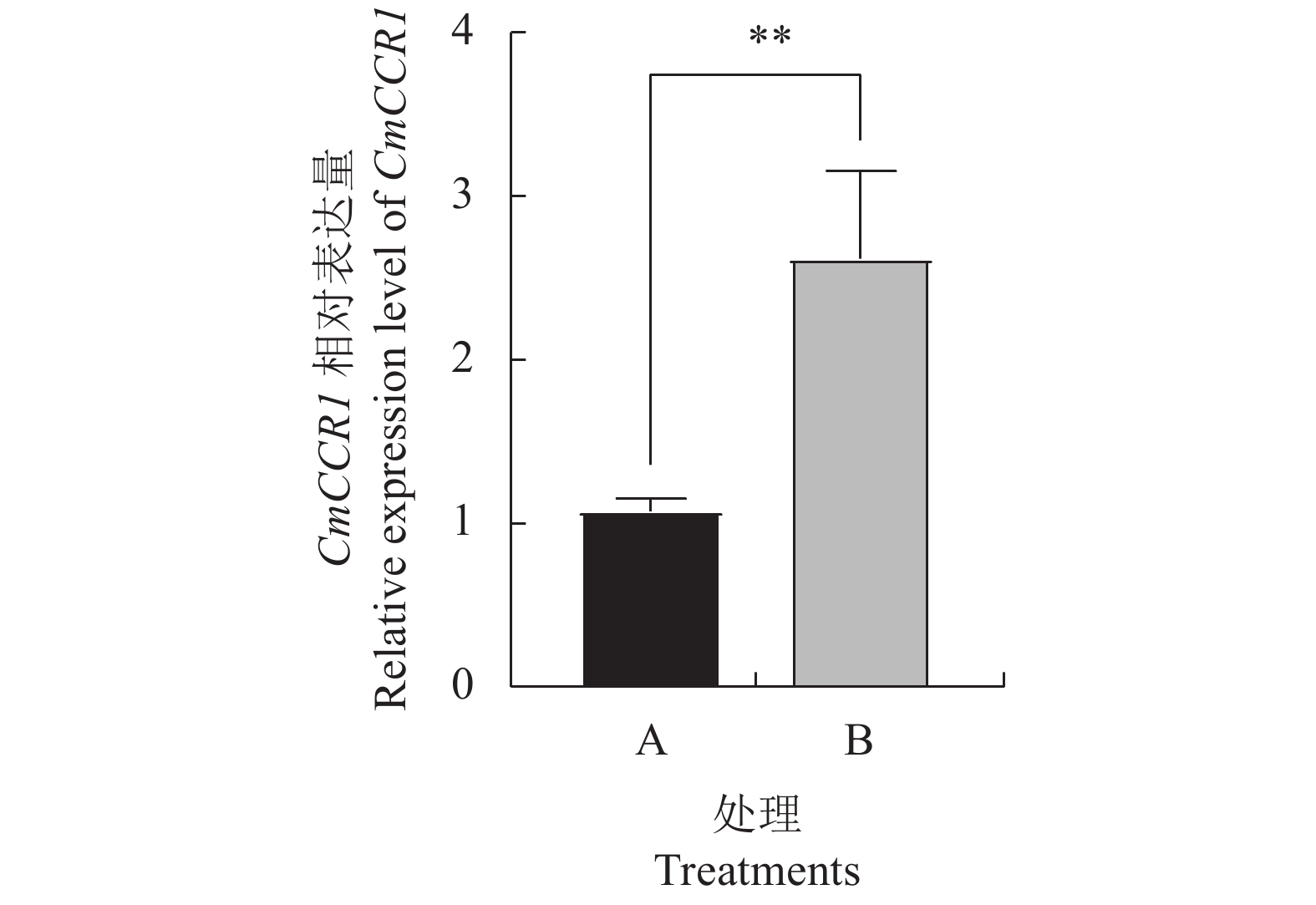
1017 bp;生物信息学分析发现,该基因编码的CmCCR1蛋白分子量为37.5 kDa,等电点(pI)7.06,属于稳定亲水蛋白,含有NAD(P)H/NAD(P)(+) 结合(NADB)域,与甜橙CcCCR1有3个相同的Motif,分布情况也大致相同;进化树分析结果表明,CmCCR1与甜橙CcCCR1的亲缘关系最近,且都归于双子叶植物分组;亚细胞定位结果表明CmCCR1定位于内质网(膜)上,与软件预测的结果相吻合;实时定量PCR结果显示CmCCR1基因的表达量中三红蜜柚汁胞成熟阶段显著上升,在六月早1号柚汁胞发育过程中的相对表达量一直处于较低水平,且显著低于同一发育阶段三红蜜柚汁胞的相对表达量;将CmCCR1瞬时转化至六月早1号柚汁胞,发现瞬时转化的汁胞中CmCCR1基因的表达量显著上升,间苯三酚染色程粉红色,说明汁胞中出现了木质素的积累。结论CmCCR1基因可能参与调控三红蜜柚汁胞木质素等次级代谢产物的生物合成,其表达量的上升与柚汁胞粒化的发生有关。
Abstract:ObjectiveFunctions and expressions of cinnamoyl-CoA reductase genes (CmCCRs) associated with the lignin biosynthesis and juice vesicle granulation in Citrus maxima were investigated.
MethodBioinformatics, gene cloning, subcellular localization, and fluorescence quantitative PCR were applied to select and clone CmCCRs from C. maxima Sanhongmiyou. Expressions of the genes in the juice sacs at different fruit development stages of Sanhongmiyou and Liuyuezaoyihao pomelos were examined. Lignin content in the sacs was determined using the acetyl bromide method.
ResultsFrom the RNA-Seq data on Sanhongmiyou juice sacs, 5 CmCCRs were identified. Since the lignin content in the mature juice sacs was shown to correlate with the gene transcription, CmCCR1 was supposed to be involved in the lignin formation, and thus further analyzed. The ORF of CmCCR1 was 1,017 bp long. The stable hydrophilic protein had a molecular weight of 37.5 kDa and a pI of 7.06 containing an NAD(P)H/NAD(P)(+) binding (NADB) domain and three shared common motifs of CcCCR1 in sweet orange with similar distribution patterns. Phylogenetically, it closely related to CcCCR1 and belonged to the dicotyledonous plants. The gene was located on the endoplasmic reticulum same as predicted previously by software. The expression of CmCCR1 in the Sanhongmiyou juice sacs increased significantly at maturation, but that of Liuyuezaoyihao remained relatively low throughout the entire developmental stages at significantly lower levels. However, when the Liuyuezaoyihao pomelo juice sacs were transiently transformed with CmCCR1, a significantly elevated expression of the gene resulted. Moreover, the phloroglucinol-stained sacs turned pink in color indicating lignin accumulation in them.
ConclusionCmCCR1 was postulated to regulate the biosynthesis of secondary metabolites (e.g., lignin) and the granulation of juice vesicles in pomelos.
-
Keywords:
- Citrus maxima /
- cinnamoyl-CoA reductase /
- gene cloning /
- expression analysis
-
0. 引言
【研究意义】胰岛素样生长因子结合蛋白2(Insulin-like growth factor binding protein 2,IGFBP2)属于IGFBPs家簇,在动物外周循环和细胞外液中广泛存在,在脂肪形成过程中发挥着十分重要的作用[1-2]。IGFBP2与胰岛素样生长因子2(IGF2)有较高的亲和力,与IGF2结合后将其转运到溶酶体降解,以调节IGF2的含量,并能抑制IGF2与MDBK细胞表面结合,进而抑制它们促有丝分裂活性功能[3]。【前人研究进展】目前,有研究报道IGFBP2基因多态性与动物生产性能、经济性状间存在关联,发现了一些影响肉质性状的SNPs与QTLs。杨华等[4]检测发现在大白猪×梅山猪杂交猪试验群体中IGFBP2基因第2内含子内存在1个C-T碱基突变,引起Msp Ⅰ酶切位点的改变,关联研究发现该多态位点与胴体性状、肉质性状间存在显著相关关系。Rohrer等[5]采用182个微卫星标记扫描了370头杜洛克猪×长白猪F2代群体的基因组,发现并证实猪15号染色体上IGBFP2基因所在区域内与大理石纹和嫩度相连锁的QTL位点可以用来提高猪肉品质。Prasongsook等[6]在美国密歇根州立大学杜洛克猪×皮特兰猪F2代资源群体中研究发现,IGFBP2基因不同基因型对失水率、pH24、肉色、多汁性和嫩度等肉质性状有显著影响。【本研究切入点】苏姜猪是以地方品种姜曲海猪为母本,导入杜洛克猪血统,运用常规育种与现代分子遗传育种手段相结合的方法培育而成的瘦肉型新品种猪,并于2013年成功通过国家畜禽遗传资源委员会审定,获得畜禽新品种证书。目前,关于苏姜猪的研究主要集中在生产性能、屠宰测定等方面[7-8],候选基因及其多态性与肉质性状等性能之间相关关系的研究报道较少。【拟解决的关键问题】为了寻找与苏姜猪肉质性状相关的遗传标记,本试验采用PCR-RFLP技术检测3个猪种IGFBP2基因Msp Ⅰ酶切片断多态性,并分析了苏姜猪不同基因型间肉质性状的差异,以期为苏姜猪持续选育及分子标记辅助选择研究提供参考依据。
1. 材料与方法
1.1 试验材料
1.1.1 试验动物
苏姜猪447头饲养于江苏苏姜种猪有限公司,姜曲海猪109头饲养于江苏姜曲海种猪场,杜洛克猪218头饲养于常州市康乐农牧有限公司。同一公司内饲养的育肥期试验猪日龄、体重相近,公母各半,饲粮营养水平相同,饲养管理方式一致。
1.1.2 主要试剂与仪器
Msp Ⅰ限制性内切酶购自赛默飞世尔科技(中国)有限公司,2×Taq PCR Mix、DL2000 DNA Marker购自北京华越洋生物科技有限公司,KAPA DNA快速提取试剂盒购自上海捷易生物科技有限公司。主要仪器包括Q5000超微量核酸蛋白测定仪、ABI Veriti 96梯度PCR仪、WSC-S测色色差计。
1.2 试验方法
1.2.1 组织样采集与DNA提取
所有试验猪在育肥结束销售前逐头采集耳组织样0.5 g,置于装有70%酒精的Eppendorf管内,冰盒保存带回实验室。按照DNA提取试剂盒说明书提取基因组DNA,采用超微量核酸蛋白测定仪检测DNA浓度和纯度,−20℃保存备用。
1.2.2 肉质性状测定
苏姜猪体重达100 kg左右时,每个基因型选取8头进行屠宰,从左半侧胴体倒数第3~4胸椎处向后采集背最长肌20~30 cm,按照“NY/T821-2004猪肌肉品质测定技术规范”测定剪切力、失水率、肉色、pH24、大理石纹等肉质性状指标。其中肉色采取5分制比色板评分法和WSC-S色差仪法2种方法测量,色差仪测的L值代表光反射值,a值和b值分别代表红度和黄度值。
1.2.3 PCR扩增
根据杨华等[4]研究成果,由生工生物工程(上海)股份有限公司合成一对引物,扩增IGFBP2基因第2内含子内245 bp片段。引物序列分别为:上游,5′-GGTCT GATTG GAGGG GTGT-3′;下游,5′-AGCCA AGGAG AAATG TGAAG G-3′。PCR 扩增反应体系包括:DNA模板2.5 μL,2×Taq PCR Mix 12.5 μL,上下游引物(10 μmol·L−1)各 1.0 μL,加ddH2O至总体积25.0 μL。PCR 反应程序:94℃预变性5 min;94℃变性45 s,56℃退火45 s,72℃延伸50 s,共计35个循环;72℃延伸10 min,并在4℃条件下保存备用。PCR扩增产物用1.5%琼脂糖凝胶电泳检测,在凝胶成像系统下观察扩增效果。
1.2.4 PCR-RFLP-Msp Ⅰ酶切分析
符合要求的PCR扩增产物用Msp Ⅰ进行酶切,酶切反应体系包括1×Buffer 1.5 μL,Msp Ⅰ限制性内切酶0.2 μL(5 U),PCR产物5 μL,加ddH2O至总体15 μL,37℃酶切反应2 h。用1.5%琼脂糖凝胶电泳检测经过限制性内切酶Msp Ⅰ酶切的产物,并凝胶成像系统下判断基因型类型。按照刘海霞等[9]研究方法计算3个猪种试验群体内的纯合度(H0)、杂合度(He)、有效等位基因数(Ne)、多态信息含量(PIC),并比较分析IGFBP2基因不同基因型对苏姜猪肉质性状的影响。
1.3 数据处理
采用SPSS 16.0 GLM对数据进行统计学分析。
2. 结果与分析
2.1 IGFBP2基因PCR扩增产物
IGFBP2基因第2内含子目标片段的PCR扩增产物电泳检测结果见图1。从图1可见PCR产物的特异性非常好,扩增片段与目标片段大小一致,长度245 bp,可以直接用来进行RFLP分析。
2.2 IGFBP2基因PCR-RFLP检测分型
对3个猪种IGFBP2基因目标片断的PCR扩增产物进行RFLP-Msp Ⅰ分析,1.5%琼脂糖凝胶电泳检测结果见图2。结果发现,IGFBP2基因PCR产物在3个猪种试验群体中均检测到了Msp Ⅰ酶切多态性,存在AA、AB和BB 3种基因型。
2.3 IGFBP2基因遗传多态性分析
2.3.1 等位基因和基因型频率
统计3个猪种试验群体IGFBP2基因目标片段的等位基因和基因型频率(表1)。由表1可知,3个猪种试验群体中优势基因型均为AB,优势等位基因为B。
表 1 IGFBP2基因基因型频率和基因频率Table 1. Genotype and frequency distribution of IGFBP2 gene品种 Breed 基因型频率 Genotype frequency 等位基因频率 Allele frequency AA AB BB A B 苏姜猪 Sujiang pig 0.16(76) 0.48(229) 0.36(172) 0.40 0.60 姜曲海猪 Jiangquhai pig 0.10(11) 0.59(64) 0.31(34) 0.39 0.61 杜洛克猪 Duroc pig 0.13(29) 0.61(132) 0.26(57) 0.44 0.56 注:括号内数字为个体数,括号外数字为频率。
Note: Number of individuals outside parentheses, frequency in parentheses.2.3.2 纯合度、杂合度、有效等位基因数及多态信息含量
3个猪种试验群体H0、He、Ne及PIC的统计结果见表2。由表2可知,3个猪种试验群体的PIC均呈现中度多态,且姜曲海猪试验群体的遗传多样性高于苏姜猪、杜洛克猪试验群体。
表 2 IGFBP2基因多态位点纯合度、杂合度、有效等位基因数和多态信息含量Table 2. Genetic polymorphism parameters ( Ho, He, Ne and PIC ) of IGFBP2 gene品种 Breed 纯合度 H0 杂合度 He 有效等位基因数 Ne 多态信息含量 PIC 苏姜猪 Sujiang pig 0.52 0.48 1.92 0.36 姜曲海猪 Jiangquhai pig 0.52 0.48 1.92 0.41 杜洛克猪 Duroc pig 0.50 0.50 2.00 0.38 注:PIC<0.25:低度多态;0.25<PIC<0.5:中度多态;PIC>0.5:高度多态。
Note: PIC<0.25: low polymorphism; 0.25<PIC<0.5: moderate polymorphism; PIC>0.5: high polymorphism.2.4 IGFBP2不同基因型对苏姜猪肉质性状的影响
苏姜猪IGFBP2基因不同基因型个体间肉质性状的关联分析见表3。由表3可知,AA、AB型个体大理石纹显著高于BB型(P<0.05),AB、BB型与AA型个体的五分制肉色和色差仪测得的a值间的差异达到显著水平(P<0.05),其余指标间的差异均不显著(P>0.05)。
表 3 苏姜猪IGFBP2基因不同基因型肉质性状间的差异分析Table 3. Comparison of meat quality of Sujiang pigs with varied IGFBP2 gene基因型 Genotype pH 失水率
Water loss rate /%大理石纹评分
Marbling score肉色评分
Meat color score肉色 Meat color L a b AA 5.57±0.04 20.94±2.04 3.83±0.45a 3.87±0.23a 43.81±0.64 15.92±0.42a 20.08±0.36 AB 5.53±0.02 23.92±0.96 3.78±0.23a 2.63±0.10b 44.17±0.54 14.01±0.37b 20.39±0.27 BB 5.61±0.05 21.83±1.25 2.34±0.18b 2.73±0.15b 44.78±0.60 13.95±0.38b 20.41±0.32 注:同列数据后不同小写字母表示差异显著(P<0.05)。
Note: Different lowercase letters in the same column represented significant difference (P<0.05).3. 讨论与结论
IGFBP2是胰岛素样生长因子(Insulin-like growth factor, IGF)家族中的一种结合蛋白,主要存在于动物组织和体液中,可参与调控细胞的生长和代谢,与细胞粘附、瘤血管形成密切相关[10]。IGFBP2的研究主要集中在人类医学方面,许多肿瘤组织都有IGFBP2表达或表达水平升高,表明IGFBP2可能是一种肿瘤的分化程度的标志物[11-12]。
前期IGFBP2基因的研究主要集中在其结构与功能方面,目前有关IGFBP2基因多态性与经济性状的关联研究逐渐增多。Zhao等[13]测序发现IGFBP2基因第2内含子区域内存在2个SNPs(C1107G、C1130T)与两个插入/缺失片段(552 bp后TC,1 115 bp后GCCAGGT),京海黄鸡试验群体中AA型个体的出雏重、12周龄体重显著高于AB型个体,A等位基因对个体体重有正面影响,并由此推测IGFBP2基因内含子2区域的突变可以用作京海黄鸡体重的遗传标记。Wang等[14]将猪IGFBP2基因定位于15号染色体(15q22-23),中国地方猪种携带的A等位基因(50%以上)比欧洲猪种要高,不同基因型个体间在前躯重、后躯重、前肢重、后肢重等生产性能间差异均达到显著或极显著水平。
本研究在苏姜猪、姜曲海猪、杜洛克猪等3个试验群体中均发现IGFBP2基因第2内含子区域内存在着1个Msp Ⅰ酶切位点多态性,获得A、B 2个等位基因,AB、AB、BB 3种基因型。苏姜猪试验群体的多态信息含量为0.36,呈现出中度多态,由此可见苏姜猪群体具备进一步选育的潜力。苏姜猪IGFBP2基因不同基因型个体在大理石纹、肉色两个指标中存在显著差异,AA型个体的大理石纹、肉色均显著高于BB型,这些结果与杨华等[4]、刘海霞等[9]的研究结论相似,但与Prasongsook等[6]在部分肉质指标间存在差异。Prasongsook等研究发现国外杂交商品猪群体中IGFBP2基因不同基因型的失水率、pH值、肉色等性状均达到了显著水平,而本试验中却没有发现苏姜猪试验群体在不同基因间的失水率、pH值存在显著差异,这可能与试验猪的品种、数量、饲养管理、测定方法等因素存在着一定的关系。由于试验条件的限制,本研究无法分析IGFBP2基因第2内含子内PCR-RFLP-Msp Ⅰ多态位点对姜曲海猪、杜洛克猪肉质性状的影响,等试验条件具备后可作进一步的分析与探讨。
-
图 1 三红蜜柚果实汁胞成熟期木质素含量与CmCCRs基因的转录水平变化
A:汁胞成熟期间木质素含量变化,B:转录组数据中的5个CCRs基因在汁胞成熟期间的表达热图,C:木质素含量和5个CCRs基因表达水平的相关性。**表示Ρ<0.01,同图10。
Figure 1. Changes on lignin content and CmCCR transcript of Sanhongmiyou juice sacs during maturation
A: changes on lignin content in juice sacs during maturation; B: transcriptome-based expression heatmap on 5 CCRs in juice sacs during maturation; C: correlation between lignin content and expressions of 5 CCRs.** indicates P<0.01, same for Fig.10.
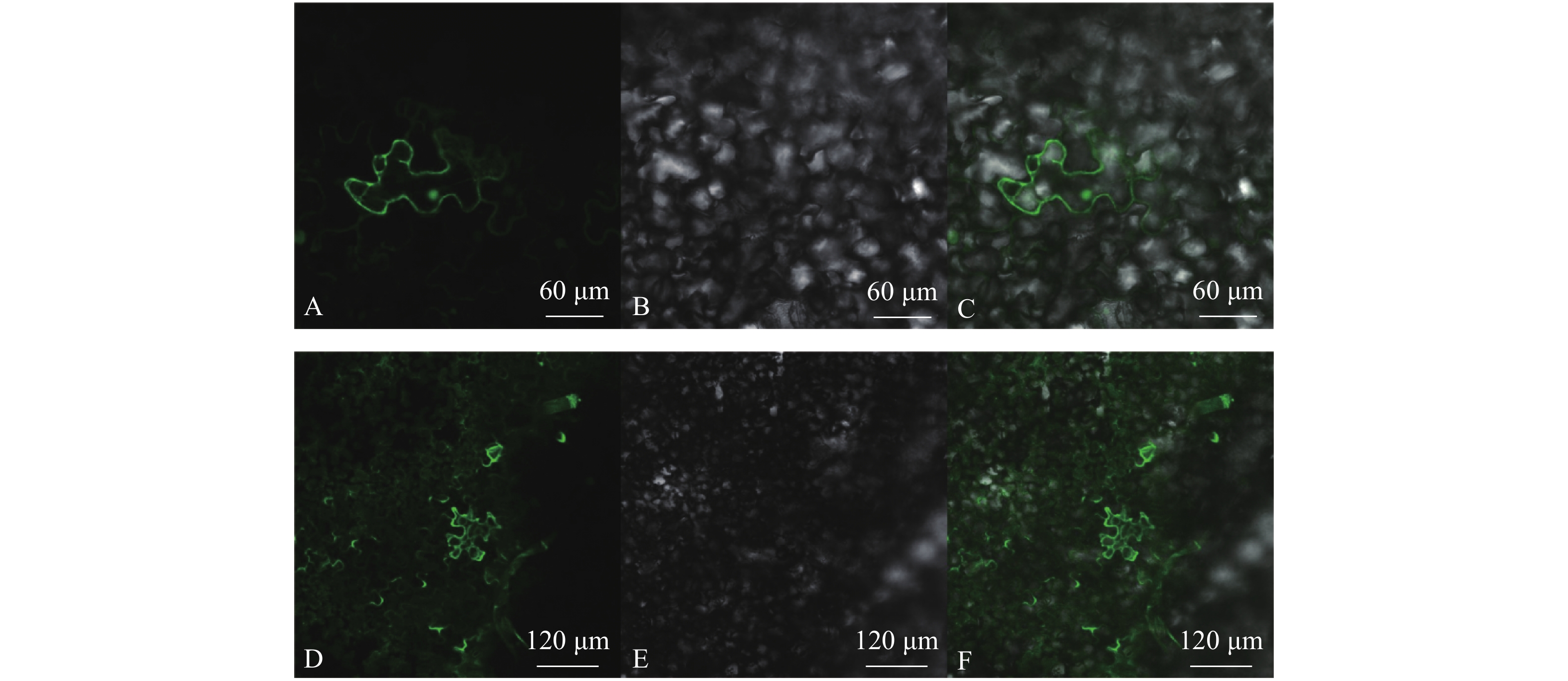
图 7 CmCCR1亚细胞定位
A~C表示p-super1300-GFP在烟草叶片中的亚细胞定位图。其中A表示p-super1300-GFP在荧光下的状态;B表示烟草叶片细胞在明场下的状态;C表示叠加效果图。D~F表示p-super1300-CmCCR-GFP在烟草叶片中的亚细胞定位图。D表示p-super1300-CmCCR-GFP在荧光下的状态;E表示烟草叶片细胞在明场下的状态;F表示叠加效果图。
Figure 7. Subcellular localization of CmCCR1 gene
A–C show subcellular localizations of p-super1300-GFP; A represents state of p-super1300-GFP under fluorescence; B represents state of tobacco leaf cells under bright field; C represents overlay effect image. D–F show subcellular localizations of p-super1300-CmCCR-GFP; D represents state of p-super1300-CmCCR-GFP; E represents state of tobacco leaf cells under bright field; F represents overlay effect image.
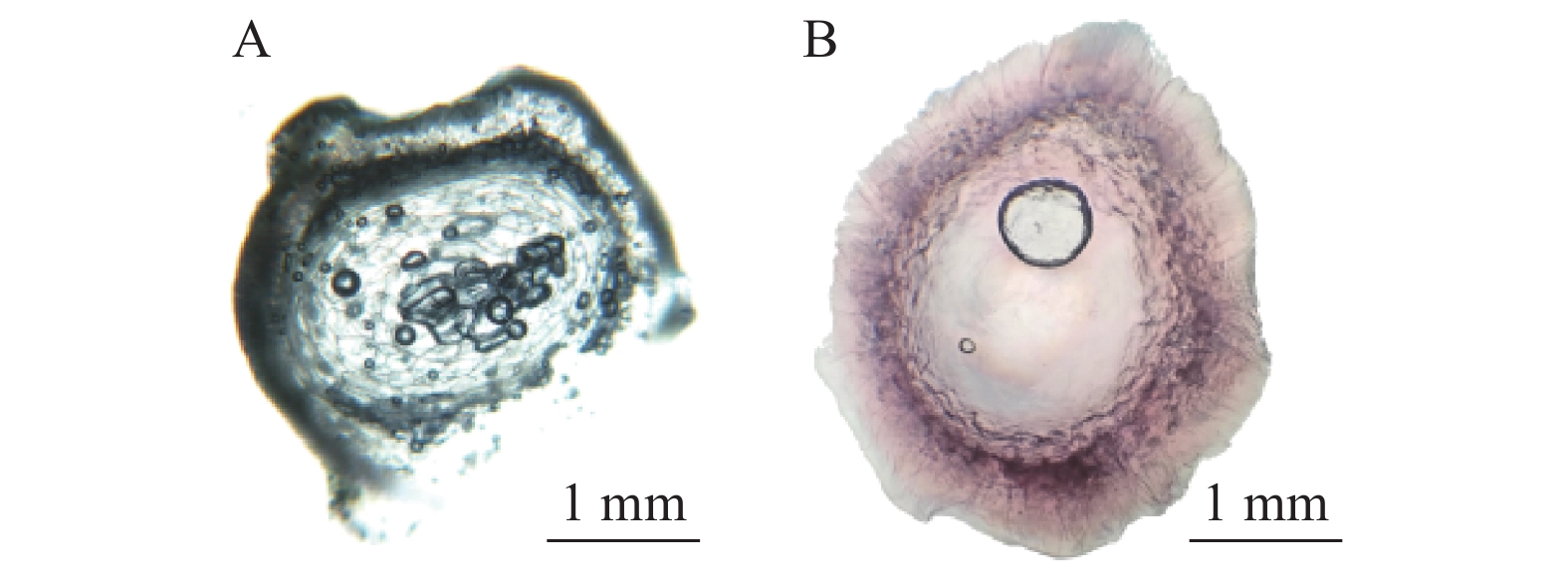
图 11 间苯三酚染色的瞬转p-CAMBIA1301-CmCCR1六月早1号柚汁胞和正常汁胞
A为对照组汁胞,未能被间苯三酚染色;B为试验组汁胞,间苯三酚染色后呈粉红色。
Figure 11. Phloroglucinol-stained juice sacs of WT and transiently transformed p-CAMBIA1301-CmCCR1 Liuyuezaoyihao
A was control group not stainable by phloroglucinol; B, treatment group of pink juice sacs stained by phloroglucinol.
表 1 CmCCR基因ORF引物序列
Table 1 Details of CmCCR ORF primers
引物
Primers引物序列
Primer sequencesCmCCR1-Xba I-F GCTCTAGAATGACGGTAATTGATAGC CmCCR1-Kpn I-R GGGGTACCGAATTTTCACTGATTC 表 2 qRT-PCR所用引物信息
Table 2 qRT-PCR primers applied
引物名称
Primer name引物序列
Primer sequencesq-CmCCR1 F: TTGATGAATCTTGCTGGAGTG R: GTCTTTGCCGAGCCATTTA Actin F: AGAACTATGAACTGCCTGATGGC R: GCTTGGAGCAAGTGCTGTGATT -
[1] 潘鹤立, 潘腾飞, 佘文琴, 等. 红绵蜜柚和三红蜜柚的园艺性状与丰产优质栽培技术 [J]. 中国南方果树, 2020, 49(4):143−148. PAN H L, PAN T F, SHE W Q, et al. Horticultural characters of hongmianmiyou and sanhongmiyou pomelos and cultivation techniques for high yield and good quality [J]. South China Fruits, 2020, 49(4): 143−148. (in Chinese)
[2] 潘东明, 郑国华, 陈桂信, 等. 琯溪蜜柚汁胞粒化原因分析 [J]. 果树科学, 1999, 16(3):202−209. PAN D M, ZHENG G H, CHEN G X, et al. Analysis of the reasons caused granulation of juice sacs in guanximiyou pomelo variety [J]. Journal of Fruit Science, 1999, 16(3): 202−209. (in Chinese)
[3] 潘腾飞, 朱学亮, 潘东明, 等. ‘琯溪蜜柚’贮藏期间汁胞粒化与木质素代谢的关系 [J]. 果树学报, 2013, 30(2):294−298. PAN T F, ZHU X L, PAN D M, et al. Relationship between granulation and lignin metabolism in ‘Guanximiyou’pummelo fruit during storage [J]. Journal of Fruit Science, 2013, 30(2): 294−298. (in Chinese)
[4] LI X T, HUANG H T, RIZWAN H M, et al. Transcriptome analysis reveals candidate lignin-related genes and transcription factors during fruit development in pomelo (Citrus maxima) [J]. Genes, 2022, 13(5): 845. DOI: 10.3390/genes13050845
[5] NEUTELINGS G. Lignin variability in plant cell walls: Contribution of new models [J]. Plant Science, 2011, 181(4): 379−386. DOI: 10.1016/j.plantsci.2011.06.012
[6] VANHOLME R, DE MEESTER B, RALPH J, et al. Lignin biosynthesis and its integration into metabolism [J]. Current Opinion in Biotechnology, 2019, 56: 230−239. DOI: 10.1016/j.copbio.2019.02.018
[7] YADAV S, CHATTOPADHYAY D. Lignin: The building block of defense responses to stress in plants [J]. Journal of Plant Growth Regulation, 2023, 42(10): 6652−6666. DOI: 10.1007/s00344-023-10926-z
[8] LEE M H, JEON H S, KIM S H, et al. Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants [J]. The EMBO Journal, 2019, 38(23): e101948. DOI: 10.15252/embj.2019101948
[9] VANHOLME R, DEMEDTS B, MORREEL K, et al. Lignin biosynthesis and structure [J]. Plant Physiology, 2010, 153(3): 895−905. DOI: 10.1104/pp.110.155119
[10] VAN ACKER R, LEPLÉ J C, AERTS D, et al. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase [J]. Proceedings of the National Academy of Sciences of the United States of America, 2014, 111(2): 845−850.
[11] LI W, HAO Z Y, YANG L C, et al. Genome-wide identification and characterization of LcCCR13 reveals its potential role in lignin biosynthesis in Liriodendron chinense [J]. Frontiers in Plant Science, 2023, 13: 1110639. DOI: 10.3389/fpls.2022.1110639
[12] LACOMBE E, HAWKINS S, VAN DOORSSELAERE J, et al. Cinnamoyl CoA reductase, the first committed enzyme of the lignin branch biosynthetic pathway: Cloning, expression and phylogenetic relationships [J]. Plant Journal, 1997, 11(3): 429−441. DOI: 10.1046/j.1365-313X.1997.11030429.x
[13] CUI W T, ZHUANG Z H, JIANG P H, et al. Characterization, expression profiling, and biochemical analyses of the Cinnamoyl-CoA reductase gene family for lignin synthesis in alfalfa plants [J]. International Journal of Molecular Sciences, 2022, 23(14): 7762. DOI: 10.3390/ijms23147762
[14] LEPLÉ J C, DAUWE R, MORREEL K, et al. Downregulation of cinnamoyl-coenzyme A reductase in poplar: Multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure [J]. The Plant Cell, 2007, 19(11): 3669−3691. DOI: 10.1105/tpc.107.054148
[15] GOUJON T, FERRET V, MILA I, et al. Down-regulation of the AtCCR1 gene in Arabidopsis thaliana: Effects on phenotype, lignins and cell wall degradability [J]. Planta, 2003, 217(2): 218−228. DOI: 10.1007/s00425-003-0987-6
[16] CHEN F, DIXON R A. Lignin modification improves fermentable sugar yields for biofuel production [J]. Nature Biotechnology, 2007, 25(7): 759−761. DOI: 10.1038/nbt1316
[17] 谢诗. 缺硼对两种柑橘砧木木质素含量及其合成关键基因表达的影响[D]. 武汉: 华中农业大学, 2014. XIE S. Effects of boron deficiency on lignin content and the key gene expression of lignin biosynthesis in two kinds citrus rootstocks[D]. Wuhan: Huazhong Agricultural University, 2014. (in Chinese)
[18] ANTEROLA A M, LEWIS N G. Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity [J]. Phytochemistry, 2002, 61(3): 221−294. DOI: 10.1016/S0031-9422(02)00211-X
[19] LIVAK K J, SCHMITTGEN T D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method [J]. Methods, 2001, 25(4): 402−408. DOI: 10.1006/meth.2001.1262
[20] FUKUSHIMA R S, KERLEY M S. Use of lignin extracted from different plant sources as standards in the spectrophotometric acetyl bromide lignin method [J]. Journal of Agricultural and Food Chemistry, 2011, 59(8): 3505−3509. DOI: 10.1021/jf104826n
[21] BARAKAT A, YASSIN N B M, PARK J S, et al. Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants [J]. Plant Science, 2011, 181(3): 249−257. DOI: 10.1016/j.plantsci.2011.05.012
[22] BARROS J, SERK H, GRANLUND I, et al. The cell biology of lignification in higher plants [J]. Annals of Botany, 2015, 115(7): 1053−1074. DOI: 10.1093/aob/mcv046
[23] LAUVERGEAT V, LACOMME C, LACOMBE E, et al. Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria [J]. Phytochemistry, 2001, 57(7): 1187−1195. DOI: 10.1016/S0031-9422(01)00053-X
[24] CHAO N, LI N, QI Q, et al. Characterization of the cinnamoyl-CoA reductase (CCR) gene family in Populus tomentosa reveals the enzymatic active sites and evolution of CCR [J]. Planta, 2017, 245(1): 61−75. DOI: 10.1007/s00425-016-2591-6
[25] MANDROU E, HEIN P R G, VILLAR E, et al. A candidate gene for lignin composition in Eucalyptus: Cinnamoyl-CoA reductase (CCR) [J]. Tree Genetics & Genomes, 2012, 8(2): 353−364.
[26] CHAO N, JIANG W T, WANG X C, et al. Novel motif is capable of determining CCR and CCR-like proteins based on the divergence of CCRs in plants [J]. Tree Physiology, 2019, 39(12): 2019−2026. DOI: 10.1093/treephys/tpz098
[27] WU D, NI M, LEI X, et al. Analyses of pepper cinnamoyl-CoA reductase gene family and cloning of CcCCR1/2 and their function identification in the formation of pungency [J]. Horticulturae, 2022, 8(6): 537. DOI: 10.3390/horticulturae8060537
[28] WANG X, ZHANG Z X, WANG W X, et al. Functional identification of CCR1 gene in apple (Malus halliana) demonstrates that it enhances saline-alkali stress tolerance [J]. Chemical and Biological Technologies in Agriculture, 2024, 11(1): 45. DOI: 10.1186/s40538-024-00565-1
[29] JONES L, ENNOS A R, TURNER S R. Cloning and characterization of irregular xylem4 (irx4): A severely lignin-deficient mutant of Arabidopsis [J]. Plant Journal, 2001, 26(2): 205−216. DOI: 10.1046/j.1365-313x.2001.01021.x
[30] YANG Z, LIU Z H, XU H, et al. ArecaceaeMDB: A comprehensive multi-omics database for Arecaceae breeding and functional genomics studies [J]. Plant Biotechnology Journal, 2023, 21(1): 11−13. DOI: 10.1111/pbi.13945
[31] YIN N W,LI B,LIU X,et al. Two types of cinnamoyl-CoA reductase function divergently in accumulation of lignins, flavonoids and glucosinolates and enhance lodging resistance in Brassica napus [J]. The Crop Journal, 2022, 10(3): 647−660.
[32] PICHON M, COURBOU I, BECKERT M, et al. Cloning and characterization of two maize cDNAs encoding cinnamoyl-CoA reductase (CCR) and differential expression of the corresponding genes [J]. Plant Molecular Biology, 1998, 38(4): 671−676. DOI: 10.1023/A:1006060101866
[33] LI J, FAN F, WANG L, et al. Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum[J]. PeerJ, 2016, 4(5): e2005.
[34] 卢晓鹏. 砂梨(Pyrus pyrifolia Nakai)果实石细胞、褐色果皮和柠檬酸形成机制研究[D]. 武汉: 华中农业大学, 2011 LU X P. Mechanism researches on the formation of stone cell, russet skin and citric acid of pear (Pyrus pyrifolia Nakai) fruit[D]. Wuhan: Huazhong Agricultural University, 2011. (in Chinese)
-
期刊类型引用(0)
其他类型引用(1)





 下载:
下载: