Identification and Sequences of Key Polyphenol-synthesis Genes in Mango Pulp
-
摘要:目的
鉴定探究杧果(Mangifera indica L.)苯丙氨酸解氨酶(phenylalanineammonialyase, PAL)与杧果多酚氧化酶(polyphenol oxidase, PPO)基因(MiPAL、MiPPO)家族成员及其在杧果果肉生长发育中的表达调控模式。
方法基于杧果基因组,利用生物信息学方法,从蛋白质特性、系统进化关系、基因结构、启动子顺式作用元件等多个方面鉴定MiPAL与MiPPO基因家族成员,并通过转录组分析鉴定MiPAL、MiPPO基因家族成员的表达模式。
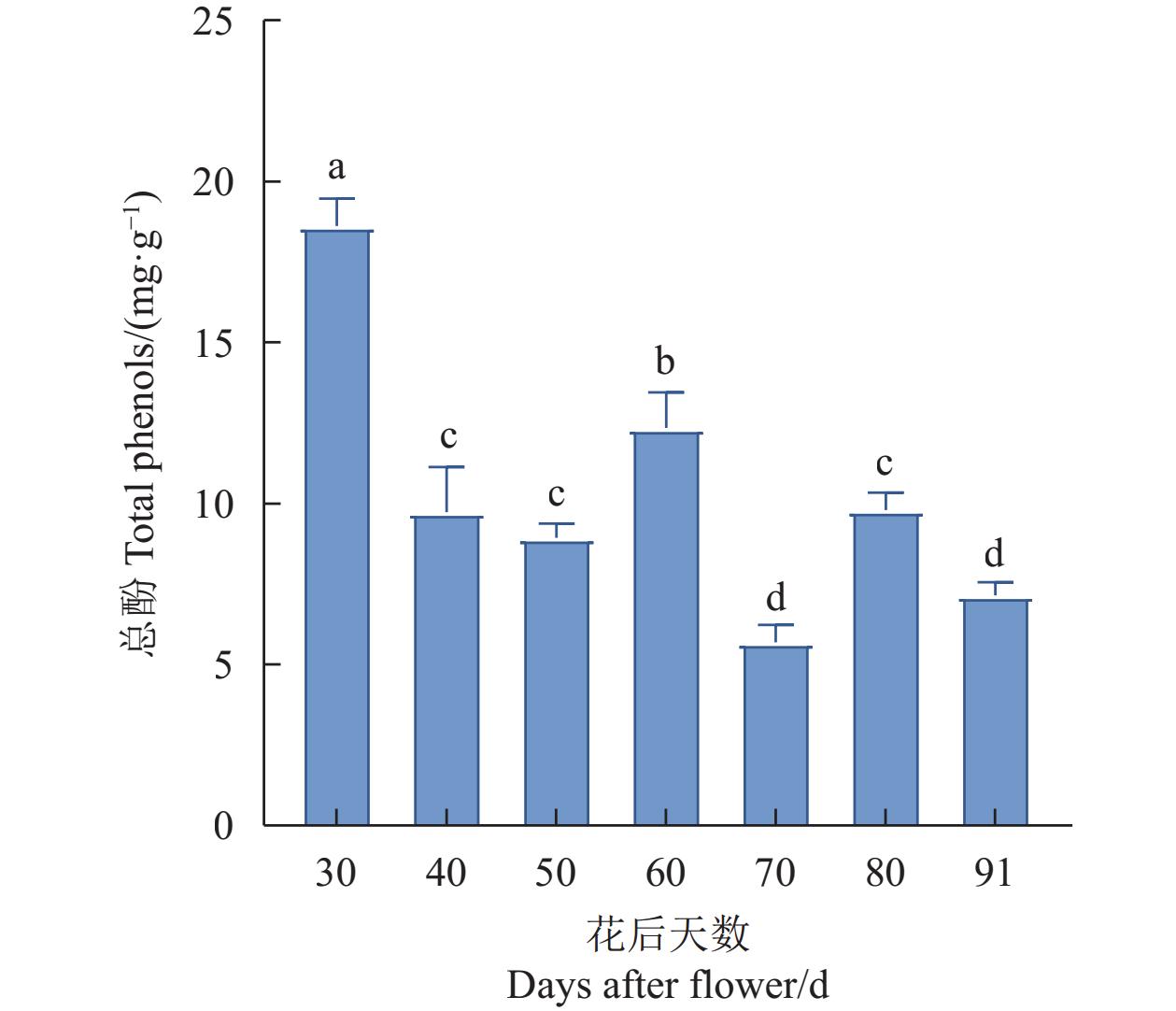
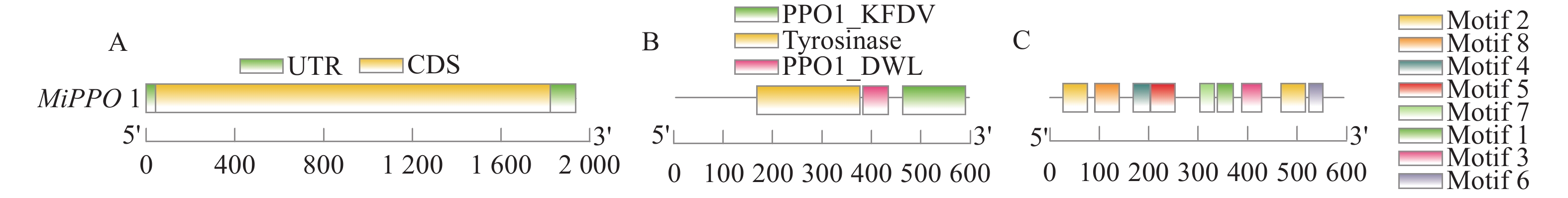
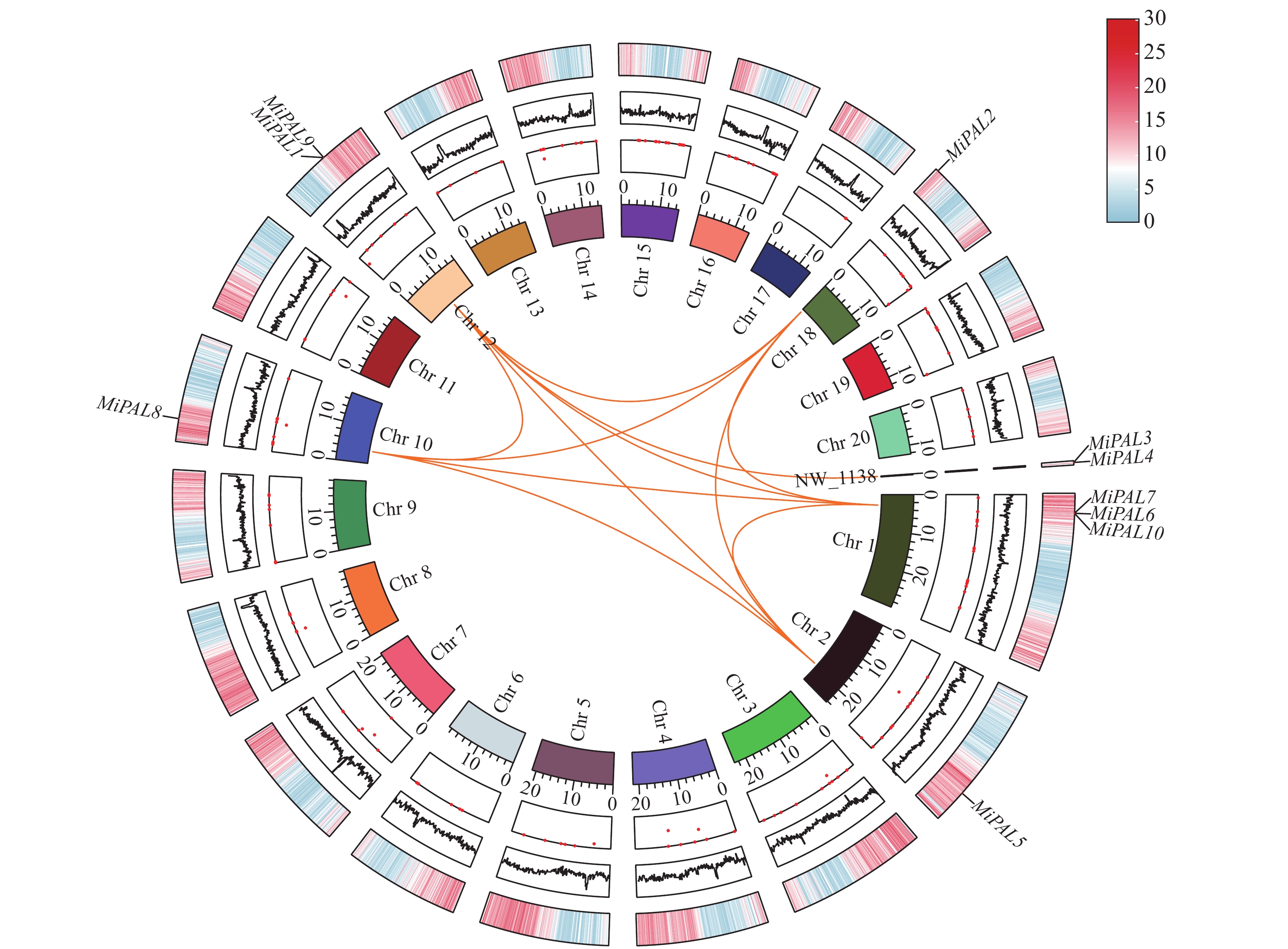
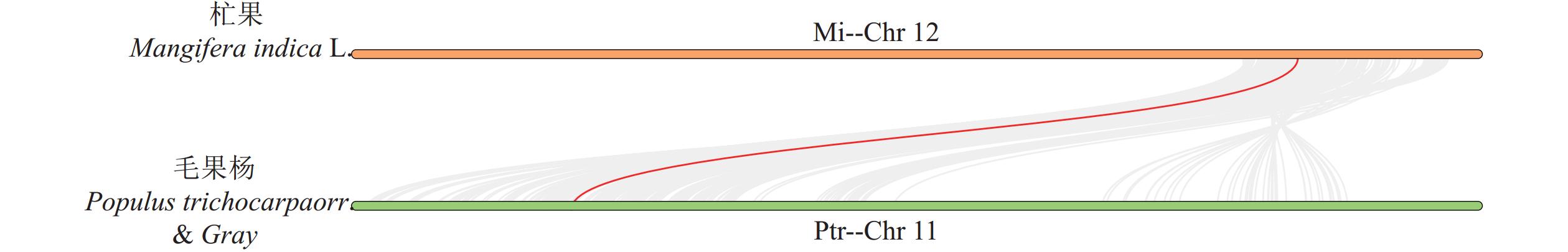
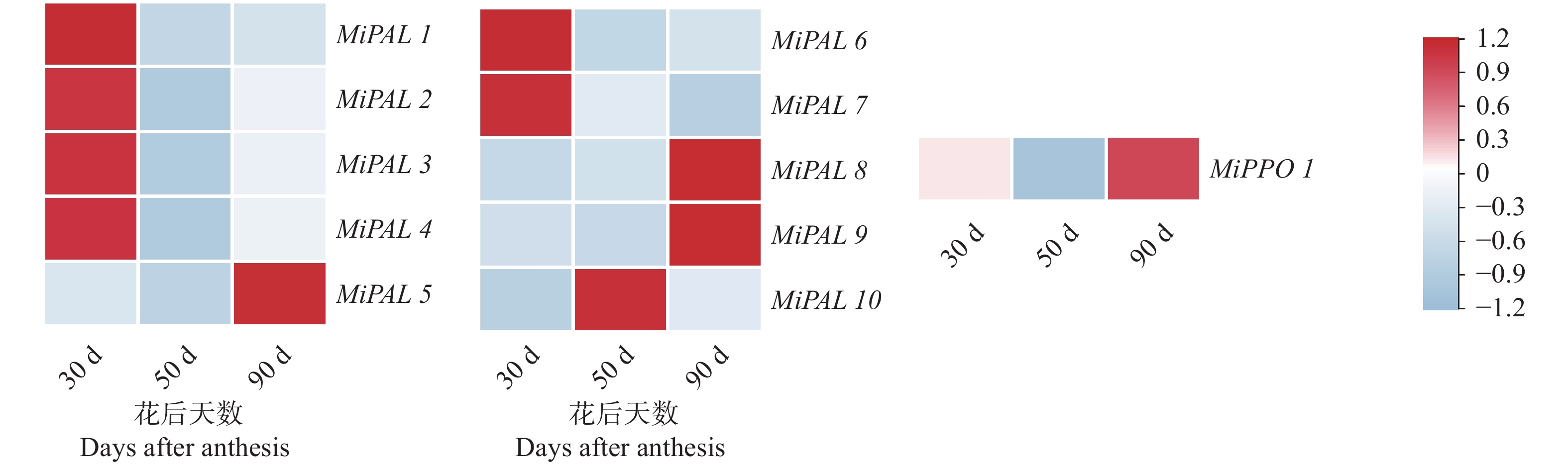
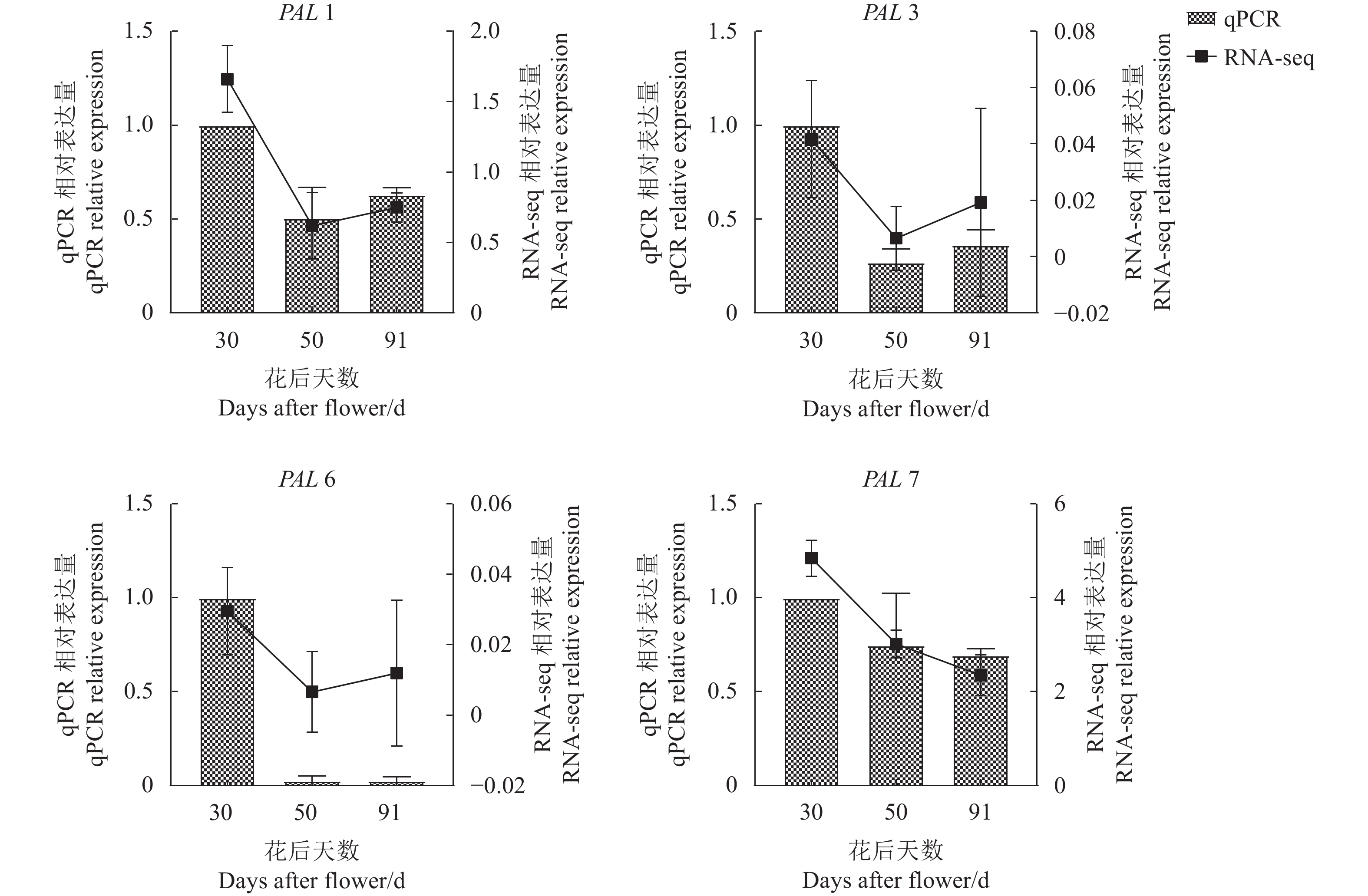
结果杧果中共鉴定出10个MiPAL基因家族成员,1个MiPPO基因家族成员;MiPAL、MiPPO蛋白均属于稳定蛋白与亲水性蛋白,多定位于叶绿体。多物种PAL、PPO基因系统发育分析表明,PAL、PPO基因均被分为4个亚组,木本植物基因家族成员多分布在同一亚组。序列分析显示,25%MiPAL基因缺少内含子,MiPPO基因无内含子序列。10个MiPAL基因分布于5条染色体与1条碎片片段上,11对片段复制基因和7个串联重复基因;MiPPO基因定位于染色体Chr12,与毛果杨(Populus trichocarpa Torr. & Gray)PPO13为同源基因。顺式作用元件分析确定了MiPAL、MiPPO成员启动子中包含生长发育元件、激素响应元件和逆境响应相关元件,表明其呈诱导型表达。MiPAL基因分别在幼果期与成熟期高表达;MiPPO基因在成熟期高表达。
结论杧果基因组中共鉴定得到10个MiPAL基因家族成员与1个MiPPO基因家族成员,同家族成员基因结构较为保守;MiPAL与MiPPO表达调控因素较多,其中MiPAL7的表达模式与杧果果肉多酚含量变化趋势相符,表明其参与了杧果果肉中多酚积累。
Abstract:ObjectiveGenes of phenylalanine ammonia lyase (MiPAL) and polyphenol oxidase (MiPPO) syntheses inMangifera indica L. were identified with their regulation functions on fruit growth and development determined.
MethodsUsing bioinformatics methods and mango genomes, the PAL and PPO family members in the fruits were identified based on analyses of protein characteristics, phylogenetic relationship, gene structure, and promoter cis acting elements. Expressions of the identified genes during fruit development were determined by transcriptome analysis.
ResultsTen PALs and onePPO in the mangos were identified. They were stable, hydrophilic proteins located mainly in the chloroplasts. Being a woody plant, these genes are phylogenetically in a same subgroup of 4 PAL and PPO categories. The sequences of 25% of MiPALs lacked introns, and that of MiPPO without intron at all. The 10 MiPALs were distributed in 5 chromosomes and one fragment with 11 pairs of segmental duplication genes and 7 tandem duplication genes. Located in chromosome Chr12, MiPPO was homologous to PPO13 in Populus tomentosa. The cis acting elements in the promoters of MiPAL and MiPPO contained those of growth, development, and hormone and stress responses expressed in an inducible pattern. MiPALs were highly expressed in the fruits at both young and mature stages, whereas MiPPO only at maturation.
ConclusionWith the information obtained, further exploration on the molecular regulatory mechanisms and biological functions of polyphenol accumulation in mango pulp was in order.
-
Keywords:
- Mangifera indica L. /
- polyphenol synthesis /
- gene family /
- gene expression
-
0. 引言
【研究意义】棕榈蓟马Thrips palmi Karny又名节瓜蓟马,属于缨翅目Thysanoptera,蓟马科Thripidae,蓟马属Thrips,分布于美国(夏威夷)、日本、印度、新加坡和中国(华南地区)等国家。主要危害茄科、葫芦科、豆科、十字花科作物,同时也是正番茄斑萎病毒属Orthotospovirus部分病毒的传播媒介[1-3]。正番茄斑萎病毒属病毒随着食物被动进入介体蓟马消化道,在消化道内建立侵染点并增殖,最终突破消化道和唾液腺屏障,病毒粒子随着唾液注入并侵染健康植株,影响植株生长、发育[4]。因此,了解消化道的组成和结构对进一步研究病毒在棕榈蓟马体内的侵染路径尤为重要。【前人研究进展】早期的缨翅目昆虫解剖学研究从形态学角度描述了缨翅目昆虫消化道、唾液腺、生殖系统等组织器官,明确不同种类蓟马的消化道形态结构存在差异[5]。此外,Ullman等利用透射电镜观察西花蓟马Frankliniella occidentalis唾液腺、前肠、中肠、后肠以及生殖系统的超薄切片,明确消化道的组织和形态结构,并探讨了蓟马消化道形态与病毒传播的关系,认为消化道形态可能决定蓟马病毒传播能力[6]。棕榈蓟马能传播番茄斑萎病毒(Tomato spotted wilt virus, TSWV)、甜瓜黄斑病毒(Melon yellow spot virus, MYSV)、花生芽枯病毒等病毒(Peanut bud necrosis virus, PBNV,)。若虫取食带毒植株5 min即可携带花生芽枯病毒,带毒成虫有效传播病毒需要取食健康植株1 h [7-10]。【本研究切入点】棕榈蓟马传播病毒的种类多,对番茄斑萎病毒、花生芽枯病毒的传播效率较高,但尚不清楚病原物在其体内的传播路径。对棕榈蓟马消化道的研究有助于更深入了解病原物的发生、复制和侵染。【拟解决的关键问题】本研究利用激光共聚焦显微镜和透射电镜在形态学和组织学上对棕榈蓟马消化道的各个部位进行研究和描述,为进一步明确病原物在消化道内的侵染路径提供依据。
1. 材料与方法
1.1 供试昆虫
棕榈蓟马于2018年4月采自云南昆明晋宁区柳坝村蔬菜大棚,鉴定后用四季豆与黄瓜饲养在人工气候箱中。饲养条件为温度27±1℃,相对湿度70%±5%,光照周期L∶D=14∶10。
1.2 消化道解剖
挑取棕榈蓟马成虫置于4℃冰箱内5~10 min,待其无活力取出放置在滴有PBS溶液(0.1 mol·L−1,pH 7.2)的载玻片上,在体视显微镜(SZ660,重庆奥特)下用镊子摘除棕榈蓟马腹部最后一节,再用镊子夹住腹部,用另外一把镊子夹住头部,将消化道从虫体中缓慢拉出,摘除多余组织。
1.3 免疫荧光样品制备
免疫荧光技术是将已知的抗原或抗体标记上荧光,再用这种荧光抗体作为探针检查细胞或组织内的相应抗原。因此,可利用该技术观察棕榈蓟马消化道组织的形态结构。将解剖出的成虫消化道迅速转移到4%多聚甲醛(Sigma)溶液中固定2 h,PBS漂洗3次后将消化道转移到2% Triton X-100(Sigma)溶液中渗透0.5 h,PBS漂洗3次后将消化道转移到含有0.5%免疫荧光染料Alexa FluorTM 633 Phalloidin(Thermo Fisher)和3%牛血清的PBS溶液中37℃孵育2 h。孵育后的消化道用PBS溶液漂洗3次,将漂洗过的消化道转移到滴有甘油的载玻片上并封片,置于Leica SP8激光共聚焦显微镜(德国Leica)下观察。
1.4 常规TEM样品制备
将解剖出的成虫消化道不同组织(前肠、中肠、后肠和马氏管)分别放入2.5%戊二醛(E-MERCK)中室温固定2 h或4℃过夜,然后用PBS溶液漂洗3次,每次15 min;再转入1%的锇酸(SPI-CHEM)溶液中固定1-2 h,PBS溶液漂洗3次,每次15 min;依次用50%、70%、80%、90%、95%的乙醇脱水,每 次15 min,最后100%乙醇和100%丙酮各脱水20 min;用spurr包埋剂(SPI-CHEM)与丙酮1∶1、1∶3分别渗透样品1 h和3 h,再用100% spurr包埋剂渗透12 h;在BJ0010聚合器(北京中兴百瑞)中70℃下聚合24 h;聚合后的样品用ULTRACUT E超薄切片机(美国AO)切片;用醋酸双氧铀(SPI-CHEM)溶液染色15 min,柠檬酸铅染色5 min,双蒸水漂洗3次,晾干后在FEI TECNAI G2透射电子显微镜(美国FEI)下观察。
2. 结果与分析
棕榈蓟马消化道始于咽部,结束于肛门,纵贯于血腔之中。根据功能不同分为前肠、中肠、后肠和马氏管(图1)。
![]() 图 1 棕榈蓟马消化道形态和结构注:Fg:前肠;Oe:食道;Cr:嗉囊;Mg:中肠;Amg:前中肠;Mmg:中中肠;Pmg:后中肠;Hg:后肠;I:回肠;Re:直肠;Mt:马氏管。Figure 1. Morphology and structure of alimentary canal of T. palmiNote: Fg: Foregut; Oe: Oesophagus; Cr: Crop; Mg: Midgut; Amg: Anterior midgut; Mmg: Middle midgut; Pmg: Posterior midgut; Hg: Hindgut; I: Ileum; Re: Rectum; Mt: Malpighian tubule.
图 1 棕榈蓟马消化道形态和结构注:Fg:前肠;Oe:食道;Cr:嗉囊;Mg:中肠;Amg:前中肠;Mmg:中中肠;Pmg:后中肠;Hg:后肠;I:回肠;Re:直肠;Mt:马氏管。Figure 1. Morphology and structure of alimentary canal of T. palmiNote: Fg: Foregut; Oe: Oesophagus; Cr: Crop; Mg: Midgut; Amg: Anterior midgut; Mmg: Middle midgut; Pmg: Posterior midgut; Hg: Hindgut; I: Ileum; Re: Rectum; Mt: Malpighian tubule.2.1 前肠(Foregut)
前肠位于消化道前端,长约300 ~350 μm,直径约15~25 μm,由咽、食道、嗉囊和贲门组成。食道较短、细,为膜质管状结构,肠腔由窄逐渐变宽,直到与嗉囊连接。嗉囊细胞和肠腔变大,肌肉组织丰富、膨大(图1)。嗉囊在与中肠连接处表皮凹陷,形成控制食物进出的圆环状贲门(图2)。通过透射电镜观察,发现前肠微绒毛多且密集,肠腔被微绒毛挤压呈缝隙状,线粒体分布在微绒毛附近,内质网靠近肠壁细胞的基膜(图3)。
2.2 中肠(Midgut)
中肠为棕榈蓟马消化道中最长的组织结构,长约900~1 000 μm,直径约40~120 μm,是消化食物和吸收营养的主要场所,可分为前中肠,中中肠和后中肠,以中肠在腹内的弯曲为分界线,前中肠最粗,直径约90~120 μm,其余粗细相似,直径约70~80 μm(图1)。前中肠肠壁由内向外分别为肠壁细胞、基膜、肌肉纤维,肠腔周围着生形状规则、细长且密集的微绒毛,微绒毛外包裹微绒毛膜(图4)。在微绒毛附近聚集大量的线粒体,形状为圆形或不规则的条形。细胞核形状不规则,在其周围分布大量粗面内质网(图4)。
后中肠与中中肠相似,直径较前中肠小,约40~55 μm,在末端内敛,与后肠连接(图1)。肠腔周围有微绒毛,无微绒毛膜,肠壁细胞在靠近微绒毛附近密集分布有线粒体。与前中肠不同的是肠壁褶皱状,肌肉镶嵌在褶皱中(图5)。
2.3 后肠(Hindgut)
后肠粗细居于前肠和中肠之间,长约300~400 μm,直径约30~40 μm,分为回肠和直肠,直肠连接肛门(图1)。回肠的形状为规则的管状,肠壁薄,只有一层扁平的肠壁细胞和一层肌肉纤维,内壁有一层致密的角质层,无微绒毛,角质层与细胞质膜之间有空隙,细胞内线粒体较少且分布无规则,肌肉发达,肌肉细胞与肠壁细胞厚度相似(图6)。直肠较回肠膨大,肠壁薄,细胞形态扁平,细胞核大,有类似微绒毛的长条突起,无角质层,肠腔内有大量微生物(图7)。
![]() 图 4 棕榈蓟马前中肠超微结构注:L:肠腔;Mv:微绒毛;Mvm:微绒毛膜;Mit:线粒体;N:细胞核;Rer:粗面内质网;In:基底膜内褶;Bm:基底膜;Mf:肌肉纤维。Figure 4. Ultrastructure of anterior midgut of T. palmiNote: L: Lumen; Mv: Microvilli; Mvm: Microvilli membrane; Mit: Mitochondria; N: Nucleus; Rer: Rough endoplasmic reticulum; In: Cuticular intima lining; Bm: Basement membrane; Mf: Muscle fiber.
图 4 棕榈蓟马前中肠超微结构注:L:肠腔;Mv:微绒毛;Mvm:微绒毛膜;Mit:线粒体;N:细胞核;Rer:粗面内质网;In:基底膜内褶;Bm:基底膜;Mf:肌肉纤维。Figure 4. Ultrastructure of anterior midgut of T. palmiNote: L: Lumen; Mv: Microvilli; Mvm: Microvilli membrane; Mit: Mitochondria; N: Nucleus; Rer: Rough endoplasmic reticulum; In: Cuticular intima lining; Bm: Basement membrane; Mf: Muscle fiber.![]() 图 5 棕榈蓟马后中肠超微结构注:①A为扁平端组织,B为褶皱端组织结构;②L:肠腔;Mv:微绒毛;Mit:线粒体;Ap:自噬体;N:细胞核;Rer:粗面内质网;Mf:肌肉纤维;Bm:基底膜。Figure 5. Ultrastructure of posterior midgut of T. palmiNote: ① A-Flat side of posterior midgut; B-Pleated side of posterior midgut; ② L: Lumen; Mv: Microvilli; Mit: Mitochondria; Ap: Autophagosome; N: Nucleus; Rer: Rough endoplasmic reticulum; Mf: Muscle fiber; Bm: Basement membrane.
图 5 棕榈蓟马后中肠超微结构注:①A为扁平端组织,B为褶皱端组织结构;②L:肠腔;Mv:微绒毛;Mit:线粒体;Ap:自噬体;N:细胞核;Rer:粗面内质网;Mf:肌肉纤维;Bm:基底膜。Figure 5. Ultrastructure of posterior midgut of T. palmiNote: ① A-Flat side of posterior midgut; B-Pleated side of posterior midgut; ② L: Lumen; Mv: Microvilli; Mit: Mitochondria; Ap: Autophagosome; N: Nucleus; Rer: Rough endoplasmic reticulum; Mf: Muscle fiber; Bm: Basement membrane.2.4 马氏管(Malpighian tubules)
马氏管着生于中肠与后肠的连接处,长约400~600 μm,直径约10~15 μm,共有4条,游离于血淋巴中,无分节或分支(图1)。管壁由单层的中空细胞构成,管腔周围有密集分布的特化的微绒毛,胞内细胞核和核仁都很大,紧挨微绒毛,线粒体大量分布于管壁细胞,中间有很多大小不一的囊泡,最外层为基膜,基膜上无肌肉纤维(图8)。
3. 讨论与结论
昆虫消化道结构形态由自身的食性和发育阶段决定。不同种类的蓟马消化道在各组织的长度、宽度和形状上存在差异[5]。蓟马类昆虫以锉吸式口器吸食植物汁液,病毒首先随植物汁液进入消化道内,随后在消化道内建立侵染点并增殖,最后突破消化道和唾液腺屏障随着唾液注入并侵染健康植株[4]。本研究结果表明,棕榈蓟马消化道结构相对简单且与已报道的其他缨翅目昆虫消化道结构大体相同,主要分为前肠、中肠、后肠和马氏管,但与已报道的西花蓟马在前肠形态和中肠超微结构上有差别。棕榈蓟马前肠的食道仅起到运输食物的作用,不能进行消化和吸收,嗉囊位于前肠食道后明显变粗的部分,与食道的区别为细胞变大,肠道直径变大,尚不明确是否具有分泌消化液、初步消化食物和吸收营养物质的功能[11]。在前肠和中肠的表皮连接处有一圈突起结构,在肠腔连接处,有一环形肌肉群为贲门,在中肠蠕动时,该结构周围肌肉紧张,会使其关闭,防止食物倒流[12]。Ullman等用扫描电镜观察了西花蓟马的前肠以及周围管状结构,在对西花蓟马前肠与中肠连接处的描述中,没有观察到前肠的外翻突起,仅在贲门处有一圈向肠腔内凹陷的瓣膜,且研究中没有提到类似嗉囊的结构[6]。
中肠是昆虫消化食物和吸收营养的主要器官。本研究表明,棕榈蓟马的中肠在腹部形成两个弯曲,以增加中肠与血淋巴的营养交换面积,使营养物质交换更充分。前中肠、中中肠与后中肠的细胞形态不同,前中肠是消化道最粗的部分,细胞大,向肠腔凸起,微绒毛密集有微绒毛膜包裹,是营养吸收的主要器官。后中肠根据细胞形态可分为扁平端和褶皱端,扁平端微绒毛稀疏,肌肉处于放松状态,褶皱端紧张时微绒毛密集。在后中肠处微绒毛未发现微绒毛膜。据此推测褶皱端细胞在肌肉放松时,细胞形态与扁平端相似,在后中肠细胞中没发现Ullman等发现的同心圆环[6]。棕榈蓟马中肠肌肉比较发达,又可分为环肌和纵肌,它们垂直交叉分布,在肠道蠕动时,原本平铺在基底膜上的肌肉会横向或者纵向收缩,与其连接的细胞形态和肠道形态也会随之改变。此外,棕榈蓟马中肠无围食膜,围食膜比较发达的昆虫的食物里微生物含量比较高,因为植物汁液中微生物较少,棕榈蓟马只进化出了一层微绒毛膜[13-14]。
大部分昆虫的回肠都是由一层角质层和不发达的肠壁细胞构成,本研究表明,棕榈蓟马回肠肌肉十分发达,几乎与肠壁细胞厚度相同[15]。此外,棕榈蓟马直肠内部还有大量的微生物,这与Ullman等在西花蓟马后肠中观察到的结果一致,在电镜下的微生物形态观察也类似[6]。
不同昆虫的马氏管形态、数量都有不同,4根马氏管是缨翅目昆虫共有的特征。本研究表明,棕榈蓟马的马氏管一端与中后肠连接处链接,一端游离在血腔中。马氏管由中空的管壁细胞构成,管壁细胞内有大量的液泡,胞内的液泡被认为是由内质网或高尔基体包裹的排泄产物[16]。
综上所述,本研究利用激光共聚焦显微镜和透射电镜解剖观察了棕榈蓟马的消化道,初步明确了棕榈蓟马消化道的构成和功能,为研究病原物在棕榈蓟马体内侵染路径提供基础依据,也可以为不同种蓟马之间传毒差异比较研究提供理论支持。
-
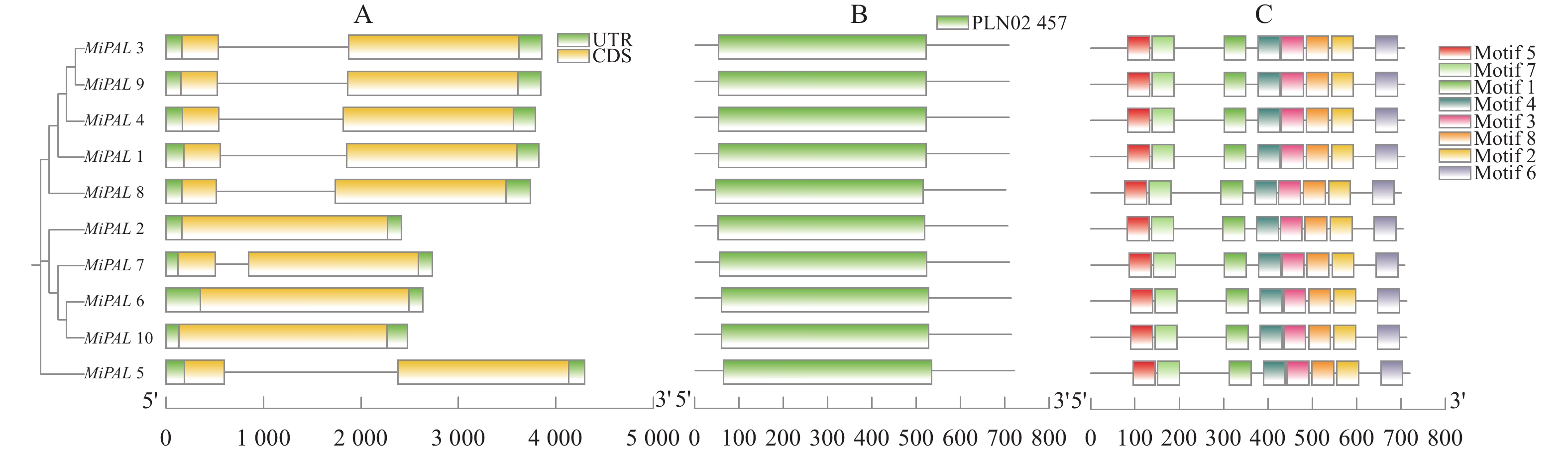
图 4 杧果PAL基因结构分析
A:基因结构分析; B:保守结构域分析; C:保守基序分析;图5同。
Figure 4. Structure of PAL in M. indica
A: Gene structure analysis; B: conservative structural domain analysis; C: conservative motif analysis. Same for Fig. 5.
表 1 杧果MiPAL及内参基因引物序列
Table 1 Sequences of primers of MiPALs and reference genes
基因名
Gene name上游引物序列 (5′-3′)
Primer sequence (5′-3′)下游引物序列 (5′-3′)
Primer sequence (5′-3′)MiPAL1 AATGGCCAACGGAGAGAGAG TGGGGACTTTACTTTCTCACCA MiPAL3 ACCAATCATGTCCAGAGCGC CCTCCAAGTGCCTCAAGTCA MiPAL6 GACCCCTGCAGTGCTACTTA ATCGCCGCATTTCCATTCTC MiPAL7 CCAATCCAGTCACCAGCCAT TGCCTCAGGTCAATCGCTTG Actin7 ATCTGCTGGAAGGTGCTGAG CCAAGCAGCATGAAGATCAA 表 2 MiPAL、MiPPO蛋白基本信息
Table 2 Basic information on MiPAL and MiPPO proteins
基因
Gene基因编号
Gene ID氨基酸数量
Number of amino acids分子量
MW/Da等电点
pI不稳定系数
Instability index脂肪系数
Aliphatic index平均亲水系数
GRAVY亚细胞定位
Subcellular localizationMiPAL1 LOC123193566.1 707 77066.14 6.47 35.46 90.11 −0.151 叶绿体 MiPAL2 LOC123201898.1 704 76781.74 6.26 32.8 91.58 −0.184 细胞质 MiPAL3 LOC123206701.1 707 76933.96 6.46 34.55 90.11 −0.144 叶绿体 MiPAL4 LOC123206728.1 707 77051.13 6.57 35.45 90.11 −0.151 叶绿体 MiPAL5 LOC123209119.1 719 78902.02 6.16 33.16 90.21 −0.229 细胞质 MiPAL6 LOC123224091.1 712 77440.26 5.76 30.42 90.7 −0.179 叶绿体 MiPAL7 LOC123225153.1 708 76938.82 6.15 30.41 92.99 −0.162 叶绿体 MiPAL8 LOC123228411.1 700 76578.56 6.06 36.46 90.6 −0.16 细胞核 MiPAL9 LOC123192215.1 707 76933.96 6.46 34.55 90.11 −0.144 叶绿体 MiPAL10 LOC123224083.1 712 77333.39 6.1 27.11 92.49 −0.163 内质网 MiPPO1 LOC123193265.1 593 66775.87 6.95 37.57 76.12 −0.501 叶绿体 表 3 杧果PAL复制基因对的Ka、Ks分析
Table 3 Ka and Ks analysis on PAL replication pairs of M. indica
同源基因
HomologsKa Ks Ka/Ks MiPAL7-MiPAL2 0.029459743 0.421135092 0.06995319 MiPAL5-MiPAL2 0.118273297 2.076835221 0.056948811 MiPAL8-MiPAL2 0.09078887 1.857097035 0.048887521 MiPAL1-MiPAL2 0.087241983 2.130415251 0.040950694 MiPAL7-MiPAL1 0.084734441 2.184662425 0.038786057 MiPAL5-MiPAL1 0.111913996 — — MiPAL8-MiPAL1 0.02388762 0.400106007 0.059703227 MiPAL1-MiPAL3 0.004306158 0.029084482 0.148056903 MiPAL7-MiPAL8 0.086248659 1.970777229 0.043763779 MiPAL5-MiPAL8 0.1112944 2.773517674 0.040127525 MiPAL7-MiPAL5 0.118588566 2.625444505 0.045168948 Ka:异义替换;Ks:用义替换。
Ka: Ambiguous substitution; Ks: meaning substitution. -
[1] IQUEBAL M A, JAISWAL S, MAHATO A K, et al. MiSNPDb: A web-based genomic resources of tropical ecology fruit mango (Mangifera indica L. ) for phylogeography and varietal differentiation [J]. Scientific Reports, 2017, 7(1): 14968. DOI: 10.1038/s41598-017-14998-2
[2] 何强, 董静雯. 果蔬中的多酚及其功能特性 [J]. 西华大学学报(自然科学版), 2019, 38(4):37−44. DOI: 10.3969/j.issn.1673-159X.2019.04.006 HE Q, DONG J W. Polyphenols in fruits and vegetables and its function [J]. Journal of Xihua University (Natural Science Edition), 2019, 38(4): 37−44. (in Chinese) DOI: 10.3969/j.issn.1673-159X.2019.04.006
[3] MUHAMMAD N, LUO Z, ZHAO X, et al. Transcriptome-wide expression analysis of MYB gene family leads to functional characterization of flavonoid biosynthesis in fruit coloration of Ziziphus Mill [J]. Frontiers in Plant Science, 2023, 14: 1171288. DOI: 10.3389/fpls.2023.1171288
[4] LI G, QIN B B, LI S D, et al. LbNR-derived nitric oxide delays Lycium fruit coloration by transcriptionally modifying flavonoid biosynthetic pathway [J]. Frontiers in Plant Science, 2020, 11: 1215. DOI: 10.3389/fpls.2020.01215
[5] ZHANG A A, ZHENG J, CHEN X M, et al. Comprehensive analysis of transcriptome and metabolome reveals the flavonoid metabolic pathway is associated with fruit peel coloration of melon [J]. Molecules, 2021, 26(9): 2830. DOI: 10.3390/molecules26092830
[6] LI S J, LI X, WANG X D, et al. Flavonoid synthesis-related genes determine the color of flower petals in Brassica napus L [J]. International Journal of Molecular Sciences, 2023, 24(7): 6472. DOI: 10.3390/ijms24076472
[7] ALVAREZ-RIVERA G, SANZ A, CIFUENTES A, et al. Flavonoid accumulation varies in Medicago truncatula in response to mercury stress [J]. Frontiers in Plant Science, 2022, 13: 933209. DOI: 10.3389/fpls.2022.933209
[8] MMBANDO G S. The recent relationship between ultraviolet-B radiation and biotic resistance in plants: A novel non-chemical strategy for managing biotic stresses [J]. Plant Signaling & Behavior, 2023, 18(1): 2191463.
[9] ZHANG Q, ZHENG G Y, WANG Q, et al. Molecular mechanisms of flavonoid accumulation in germinating common bean (Phaseolus vulgaris) under salt stress [J]. Frontiers in Nutrition, 2022, 9: 928805. DOI: 10.3389/fnut.2022.928805
[10] WANG J, GAO X Q, WANG X, et al. Exogenous melatonin ameliorates drought stress in Agropyron mongolicum by regulating flavonoid biosynthesis and carbohydrate metabolism [J]. Frontiers in Plant Science, 2022, 13: 1051165. DOI: 10.3389/fpls.2022.1051165
[11] GOURLAY G, MA D W, SCHMIDT A, et al. MYB134-RNAi poplar plants show reduced tannin synthesis in leaves but not roots, and increased susceptibility to oxidative stress [J]. Journal of Experimental Botany, 2020, 71(20): 6601−6611. DOI: 10.1093/jxb/eraa371
[12] DIXIT G, SRIVASTAVA A, RAI K M, et al. Distinct defensive activity of phenolics and phenylpropanoid pathway genes in different cotton varieties toward chewing pests [J]. Plant Signaling & Behavior, 2020, 15(5): 1747689.
[13] CHEN Q, LIANG X, WU C L, et al. Overexpression of leucoanthocyanidin reductase or anthocyanidin reductase elevates tannins content and confers cassava resistance to two-spotted spider mite [J]. Frontiers in Plant Science, 2022, 13: 994866. DOI: 10.3389/fpls.2022.994866
[14] 朱三明, 郑敏敏, 田恬, 等. 植物次生代谢途径与调控研究进展 [J]. 植物生理学报, 2023(12):2188−2216. ZHU S M, ZHEN M M, TIAN T, et al. Research progress on plant secondary metabolism and regulation [J]. Plant Physiology Journal, 2023(12): 2188−2216.
[15] 高媛, 马帅, 代敏, 等. 果蔬酚酸生物合成及代谢调控研究进展 [J]. 食品科学, 2018, 39(9):286−293. DOI: 10.7506/spkx1002-6630-201809043 GAO Y, MA S, DAI M, et al. Progress in research on the biosynthesis pathway and metabolic regulation of phenolic acids [J]. Food Science, 2018, 39(9): 286−293. (in Chinese) DOI: 10.7506/spkx1002-6630-201809043
[16] BARROS J, DIXON R A. Plant phenylalanine/tyrosine ammonia-lyases [J]. Trends in Plant Science, 2020, 25(1): 66−79. DOI: 10.1016/j.tplants.2019.09.011
[17] LIU W X, FENG Y, YU S H, et al. The flavonoid biosynthesis network in plants [J]. International Journal of Molecular Sciences, 2021, 22(23): 12824. DOI: 10.3390/ijms222312824
[18] 甄文娜. UV-B处理促进鲜切胡萝卜酚类物质富集的调控机理研究[D]. 山东: 齐鲁工业大学, 2024. ZHEN W N. Study on the regulation mechanism of UV-B treatment promoting the enrichment of phenols in fresh-cut carrots[D]. Shandong: Qilu University of Technology, 2024.
[19] ZHANG S W, QI X L, ZHU R Y, et al. Transcriptome analysis of Salvia miltiorrhiza under drought stress [J]. Plants, 2024, 13(2): 161. DOI: 10.3390/plants13020161
[20] JIANG S, WENG B S, LIU T, et al. Response of phenolic metabolism to cadmium and phenanthrene and its influence on pollutant translocations in the mangrove plant Aegiceras corniculatum (L. ) Blanco (Ac) [J]. Ecotoxicology and Environmental Safety, 2017, 141: 290−297. DOI: 10.1016/j.ecoenv.2017.03.041
[21] JAŃCZAK-PIENIĄŻEK M, CICHOŃSKI J, MICHALIK P, et al. Effect of heavy metal stress on phenolic compounds accumulation in winter wheat plants [J]. Molecules, 2022, 28(1): 241. DOI: 10.3390/molecules28010241
[22] Bibhuti Bhusan Mishra Bibhuti x. Polyphonel oxidases: Biochemical and molecular characterization, distribution, role and its control [J]. Enzyme Engineering, 2016, 5(1): 141−149.
[23] GANDÍA-HERRERO F, GARCÍA-CARMONA F. Biosynthesis of betalains: Yellow and violet plant pigments [J]. Trends in Plant Science, 2013, 18(6): 334−343. DOI: 10.1016/j.tplants.2013.01.003
[24] ONO E, HATAYAMA M, ISONO Y, et al. Localization of a flavonoid biosynthetic polyphenol oxidase in vacuoles [J]. Plant Journal, 2006, 45(2): 133−143. DOI: 10.1111/j.1365-313X.2005.02625.x
[25] LIU H, HE Q G, HU Y Y, et al. Genome-wide identification and expression profile analysis of the phenylalanine ammonia-lyase gene family in Hevea brasiliensis [J]. International Journal of Molecular Sciences, 2024, 25(9): 5052. DOI: 10.3390/ijms25095052
[26] ZHANG F L, WANG J, LI X G, et al. Genome-wide identification and expression analyses of phenylalanine ammonia-lyase gene family members from tomato (Solanum lycopersicum) reveal their role in root-knot nematode infection [J]. Frontiers in Plant Science, 2023, 14: 1204990. DOI: 10.3389/fpls.2023.1204990
[27] AMJAD M, WANG Y X, HAN S M, et al. Genome wide identification of phenylalanine ammonia-lyase (PAL) gene family in Cucumis sativus (cucumber) against abiotic stress [J]. BMC Genomic Data, 2024, 25(1): 76. DOI: 10.1186/s12863-024-01259-1
[28] GAO X G, HU Y P, XU Z B, et al. Expression profiling of the phenylalanine ammonia-lyase (PAL) gene family in ginkgo biloba L [J]. Plant Signaling & Behavior, 2023, 18(1): 2271807.
[29] ZHANG H Y, ZHANG X H, ZHAO H X, et al. Genome-wide identification and expression analysis of phenylalanine ammonia-lyase (PAL) family in rapeseed (Brassica napus L. ) [J]. BMC Plant Biology, 2023, 23(1): 481. DOI: 10.1186/s12870-023-04472-9
[30] RAWAL H C, SINGH N K, SHARMA T R. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants[J]. International Journal of Genomics, 2013, 2013: 678969.
[31] TRAN L T, TAYLOR J S, CONSTABEL C P. The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion [J]. BMC Genomics, 2012, 13: 395. DOI: 10.1186/1471-2164-13-395
[32] HE F, SHI Y J, ZHAO Q, et al. Genome-wide investigation and expression profiling of polyphenol oxidase (PPO) family genes uncover likely functions in organ development and stress responses in Populus trichocarpa [J]. BMC Genomics, 2021, 22(1): 731. DOI: 10.1186/s12864-021-08028-9
-
期刊类型引用(2)
1. 王谅,刘宇艳,李恒,陈艺欣,林硕,余芸,田厚军,林涛,张洁,陈勇,魏辉. 豆大蓟马消化道形态及超微结构. 昆虫学报. 2022(02): 144-156 .  百度学术
百度学术
2. 王小奇,梁舒勍,鲁莹. 6种蝗虫消化道的超微结构与食性相关性研究. 信阳师范学院学报(自然科学版). 2020(04): 553-559 .  百度学术
百度学术
其他类型引用(1)





 下载:
下载: