Transcriptome Changes of Citrus grandis Seedlings in Response to Acid Rain Stress
-
摘要:目的 研究琯溪蜜柚受酸雨胁迫的内在分子机制,为琯溪蜜柚科学种植提供基础资料,也为酸雨逆境生理提供理论基础。方法 以模拟酸雨胁迫24 h的琯溪蜜柚叶片进行Illumina HiSeq TM 4000 高通量转录组测序分析,将组装得到的基因在参考基因组、Nr和 KEGG 数据库进行比对;利用 FDR 与 log2(FC)来筛选差异基因,筛选条件为 FDR<0.05且|log2(FC)|>1,将筛选的差异基因做GO和KEGG富集分析。结果 模拟酸雨喷淋24 h后,琯溪蜜柚嫩叶出现明显块状伤斑;共得到21497个基因,全部得到注释,与对照相比,酸雨处理组中有879个基因显著上调,588个基因显著下调;筛选出样本中前50个DEGs(差异基因),均为上调表达基因,大部分涉及代谢途径、次生代谢、苯丙基类丙烷生物合成和萜类物质合成等相关基因。GO富集分析表明差异表达基因主要位于细胞外区域;执行分子功能中催化剂活性是最为显著富集的GO term,其次是氧化还原酶活性;生物过程中差异表达基因最显著的GO term是DNA 代谢过程。KEGG富集分析表明DNA复制是差异表达基因中最显著富集的Pathway,其次是次生代谢的生物合成,再次是苯丙素类合成途径。对次生代谢合成途径中4个差异表达基因[POD同工酶cg3g018770和cg2g001440、肉桂酰辅酶A还原酶(CCR)同工酶cg1g021310、4-香豆酸-辅酶a连接酶(4CL)同工酶cg3g029290]进行PCR荧光定量分析,验证了转录组数据的可靠性。结论 琯溪蜜柚对酸雨耐性较强,对酸雨胁迫的响应是多基因参与、多生物过程协同调控的过程,次生代谢的调节可能是应对酸雨胁迫的主要方式。Abstract:Objective Molecular mechanisms of Citrus grandis (L.) Osbeck. cv. Guanximiyou in response to simulated acid rain stress were investigated.Method The llumina HiSeq 4000 system, a high-throughput transcriptome sequencing technology, was applied to reveal the differential expressions of the grapefruit transcriptome after a 24 h simulated acid rain treatment. The unigenes obtained were compared to the Nr and KEGG database. Abundance of gene expression of the samples were screened according to transcriptome data by using PRKM method. Differentially expressed genes (DEGs) among the treated samples were estimated by referring to the standard of FDR ≤ 0.05 and |log2FC| ≥ 1. Functions and pathways of those DEGs were analyzed using the Gene Ontology (GO) and KEGG pathway database.Result Significant lumpy lesions began to appear on the young grapefruit leaves 24 h after the artificial acid rain spray with 21 497 fully described unigenes obtained. In comparison to control, 879 of the genes were significantly upregulated and 588 downregulated. The top 50 DEGs were all in the upregulated category and mainly associated with metabolic pathways, secondary metabolism, phenylpropyl propane biosynthesis, or terpenoid biosynthesis. The GO enrichment analysis showed the DEGs being largely located in extracellular region and, among various molecular functions, the catalytic activity being the most significantly enriched, followed by oxidoreductase activity, while the DNA metabolism being the most significant of DEGs in the GO term on biological process. The KEGG enrichment analysis indicated that DNA replication was the most significant enrichment pathway, followed by secondary metabolic biosynthesis, and lignin synthesis. The PCR fluorescence quantitative analysis on the 4 DEGs in the secondary metabolic biosynthesis pathway [i.e., POD isozyme cg3g018770 and cg2g001440, cinnamyl coenzyme A reductase (CCR) isozyme cg1g021310, and 4-coumaric acid-coenzyme A ligase (4CL) isozyme cg3g029290] confirmed that the acid rain stress indeed significantly affected the expressions of these genes.Conclusion C. grandis (L.) Osbeck. cv. Guanximiyou was strongly tolerant to acid rain. The stress response of the plants involved numerous genes regulated by various collaborative biological processes. Among them, the regulation of secondary metabolism appeared to play a major role in coping with acid rain stress by the plant.
-
0. 引言
【研究意义】琯溪蜜柚Citrus grandis (L.) Osbeck. cv. Guanximiyou是我国优质栽培名柚,为芸香科柑橘属果树,其果大皮薄、汁多柔软、香甜微酸,原产于福建省漳州市平和县,已有500多年栽培历史[1-2]。近些年来平和蜜柚种植面积和产量逐年增加,平和被冠以“世界柚乡,中国柚都”之称,是我国最大柚类生产和出口基地。平和县位于福建省漳州市,属南亚热带气候,琯溪蜜柚果园土壤为南亚热带较为典型的酸性红壤,由于多年来酸沉降和农业生产活动造成土壤严重酸化,是蜜柚产业面临的主要危机[3-5]。【前人研究进展】酸雨可影响植物叶片结构,破坏植物角质层,使得酸性成分通过气孔或表皮扩散进入细胞,改变植物茎叶细胞质酸碱平衡,增加细胞膜透性使得叶片中钾、钙、镁等营养元素大量淋失,影响参与基础代谢、细胞建成、光合和转录等过程的蛋白表达水平[6-8]。如酸雨处理显著上调了拟南芥水杨酸信号转导途径和茉莉酸信号通路的基因,也引起拟南芥体内基础代谢、其他信号转导途径相关基因发生变化[9],使得植株对病原菌具有更强的耐受性[10]。牛力[11]研究发现拟南芥有37个与植物次生代谢相关基因表达量发生变化,木质素、生物碱等次生代谢产物的合成在酸雨处理下加强。张琼等[12]研究发现虽然琯溪蜜柚叶片具有较强的酸雨抗性,但重度酸雨胁迫(pH 2.5)显著抑制叶肉栅栏组织发育和叶片光合速率。【本研究切入点】植物的耐酸机理是个复杂过程,琯溪蜜柚种植区位于酸雨分布区内,虽然前期已经报道了酸雨对琯溪蜜柚理化性质的影响[12],但酸雨胁迫对琯溪蜜柚影响的分子机制还鲜见报道。因此对酸雨胁迫下琯溪蜜柚叶片转录组分析可以从分子生物学层面上阐明琯溪蜜柚耐酸机制,为酸雨区琯溪蜜柚种植提供基础资料。【拟解决的关键问题】本研究以2年生琯溪蜜柚植株为试验材料,通过Illumina测序平台,比较转录组水平上模拟酸雨处理(pH 2.5)对幼嫩叶片的影响,分析可能参与抗酸或耐酸调控的相关基因,进一步揭示琯溪蜜柚受酸雨胁迫的响应机理,为琯溪蜜柚科学种植和可持续发展提供理论支持,也为酸雨逆境生理提供基础资料。
1. 材料与方法
1.1 试验材料的培育与处理
春天天气回暖,琯溪蜜柚萌发较快,2018年在2月初修剪2年生琯溪蜜柚,3月初长出部分嫩叶时,选出6棵长势一致的植株,随机分为2组,于上午10:00左右对植株进行模拟pH为2.5的酸雨3个重复喷淋(标记为T-1、T-2、T-3),同时清水3个重复喷淋为对照(标记为CK-1、CK-2、CK-3),处理使得嫩叶上下布满水滴,喷淋24 h后采集至液氮,后送至−80 ℃冰箱冷冻保存备用。
1.2 总RNA提取、cDNA合成和转录组测序
将冷冻保存的琯溪蜜柚叶片置于干冰盒中,送至广州基迪奥生物科技有限公司提取总RNA,经过磁珠[带有Oligo(dT)]富集纯化mRNA。将得到的mRNA随机打断为短片段,以这些短片段mRNA为模板,合成cDNA,通过纯化、洗脱、修复、回收,最后进行PCR扩增,完成文库制备。用Illumina HiSeqTM对文库进行高通量测序,整个流程由广州基迪奥生物科技有限公司完成。
1.3 比对参考基因组和转录本组装合并
将过滤完核糖体的 reads利用比对软件TopHat2[13]比对到参考基因组[citrus.hzau.edu.cn Genome of Pummelo (Citrus grandis)]上,再用Cufflinks[14]进行组装。将多个样品的组装结果用 Cuffmerge 进行合并,过滤错误的转录本后生成唯一注释文件。
1.4 差异表达基因筛选和功能注释分析
以log2|Fold Change| > 1 & FDR < 0.05为标准筛选出样本间差异表达基因,以未进行酸雨喷淋处理的CK为对照处理,统计酸雨处理样本表达基因的上下调关系。利用GO和KEGG数据库对筛选的差异基因进行分析。将差异表达基因在GO数据库(http://www.geneontology.org/)各条目进行映射,计算相应条目的基因数,可得条目基因列表和基因数目统计。再通过超几何检验,筛选出差异表达基因中显著富集的GO条目,假设检验的p-value计算公式是:
P=1−m−1∑i=0(Mi)(N−Mn−i)(Nn) 公式中,N是所有Unigene中具有GO注释的基因数目,n是N中差异表达基因数目,M是所有Unigene中注释为某特定GO条目的基因数目,m是注释为某GO条目的差异表达基因数目。由公式计算的值通过FDR校正后,当Q值≤0.05时,认为是差异表达基因中显著富集的GO条目。
同样通过超几何检验,以KEGG Pathway为单位进行Pathway显著性富集分析。
1.5 次生代谢相关酶PCR分析
选择4个与次生代谢相关的差异显著基因[POD同工酶cg3g018770和cg2g001440、肉桂酰辅酶A还原酶(CCR)同工酶cg1g021310、4-香豆酸-辅酶a连接酶(4CL)同工酶cg3g029290]进行验证,内参基因选用Cg3g024100(tubulin beta-6 chain)参考王莉嬛等的研究[2],反应体系按照表1配制,引物序列见表2。PCR反应条件:95 ℃ 90 s,95 ℃ 5 s,60 ℃ 15 s,72 ℃ 20 s,40 次循环。
表 1 qPCR 反应体系Table 1. qPCR reaction systemqPCR反应体系
qPCR reaction system体积
Volume/μlqPCR反应混合物 qPCR Master Mix 10 PCR正向引物PCR Forward Primer/(10 mol·L−1) 0.4 PCR反向引物 PCR Reverse Primer/(10 mol·L−1) 0.4 cDNA模板 cDNA template 4 ddH2O 5.2 合计 Total 20 表 2 引物序列Table 2. Primer sequence基因
Gene序列
SequenceCg3g024100 F 5′CAATGTGAAGTCCAGCGTGTG3′ R 5′AGCCCTCGTAGTTCTCGTCA3′ Cg3g018770 F 5′CATTTACCAATCGCCTCTATCC3′ R 5′ACCCCTGTCGGTTCATCAAGT3′ Cg2g001440 F 5′GGCAATCCAGACCCAACACT3′ R 5′AATGGCAGCGGTATCGGC3′ Cg1g021310 F 5′GTTGACGAACATTGCTGGAGT3′ R 5′GCAGCAATGTCCCTATCACC3′ Cg3g029290 F 5′CGAAGTAGAGTCCCTCAAGCA3′ R 5′TGTCACCAGTATGAAGCCAACC3′ 1.6 数据统计与分析
转录组数据在信息分析前对原始数据进行质控,并对数据过滤;按照 edgeR 的一般过滤标准(log2|Fold Change|>1 且 FDR<0.05)筛选差异基因;PCR定量分析数值为3次重复平均值,数据作图采用Origin 8.0软件。
2. 结果与分析
2.1 叶片伤害表型
由图1可知模拟酸雨喷淋24 h后,琯溪蜜柚嫩叶出现明显块状伤斑,而对照清水喷淋后嫩叶没有出现伤斑,表明pH 2.5的模拟酸雨破坏叶表面角质层,损害叶片表皮结构。
2.2 转录组质量检测和RAW数据整理分析
由表3可知通过对照组和处理组琯溪蜜柚叶片样品的转录测序,获得7 361 352 600~9 249 897 000有效数据。对数据进行质控、过滤得到7 111 541 211~8 926 570 988数据,其Q30碱基百分比均不小于95.19%,GC含量均高于44%,表明测序结果可靠,可用于后续分析。
表 3 有效数据评估统计Table 3. Statistics of valid data样本
Sample处理后
序列数据
Clean
Data/bp过滤后的
序列数据
HQ Clean
Data/bp处理后
序列数
Clean Reads
Num/条过滤后的序列
HQ Clean ReadsQ20 Q30 GC 数量
Number/条占比
Ratio/%数量
Number/bp占比
Ratio/%数量
Number/bp占比
Ratio/%数量
Number/bp占比
Ratio/ %CK-1 7361352600 7111541211 49075684 48127600 98.07 6999438082 98.42 6769749153 95.19 3153041858 44.34 CK-2 8465603100 8202153477 56437354 55469140 98.28 8080142163 98.51 7826038154 95.41 3620789302 44.14 CK-3 8683275600 8408343801 57888504 56867832 98.24 8281909562 98.5 8019114646 95.37 3726434942 44.32 T-1 8203212900 7937070962 54688086 53695546 98.19 7816272312 98.48 7566409577 95.33 3522622180 44.38 T-2 9249897000 8926570988 61665980 60549386 98.19 8792579583 98.5 8514792020 95.39 3962645890 44.39 T-3 7880490300 7621577079 52536602 51567848 98.16 7503238757 98.45 7259492038 95.25 3369836120 44.21 注:1.Q20-碱基质量 ≥20的碱基;2.Q30-碱基质量 ≥30的碱基;3.GC-鸟嘌呤和胞嘧啶;4.CK-1、CK-2、CK-3为清水对照3个重复,T-1、T-2、T-3为pH2.5模拟酸雨喷淋的3个重复,表4、5同,图2、4同。
Note: 1. Q20- Base quality≥20. 2. Q30- Base quality≥30.3. GC- Guanine and Cytosine.4.CK-1, ck-2 and ck-3 are three repetitions of clear water control, and T-1, T-2 and T-3 are three repetitions of pH2.5 simulated acid rain spray.The same as Tab. 4 and. The same as Fig.2 and 4.使用短 reads 比对工具 bowtie将过滤后的数据比对到核糖体数据库中,将比对上核糖体的数据去除,剩下的数据用于转录组的组装和分析。未比对上rRNA的reads数量是47 756 642~59 953 992,比例在98.95%~99.36%(表4)。
表 4 过滤后的序列与核糖体RNA 的比对统计Table 4. Statistics of HQ-clean data and rRNA样品
Sample过滤后的序列数
HQ Clean Reads
Number/条比对上rRNA的序列
Mapped Reads未比对上rRNA的序列
Unmapped Reads数量
Number/条占比
Ratio/%数量
Number/条占比
Ratio/%CK-1 48127600 370958 0.77 47756642 99.23 CK-2 55469140 435276 0.78 55033864 99.22 CK-3 56867832 597704 1.05 56270128 98.95 T-1 53695546 404974 0.75 53290572 99.25 T-2 60549386 595394 0.98 59953992 99.02 T-3 51567848 327954 0.64 51239894 99.36 将过滤完核糖体的数据用转录组数据比对软件TopHat2比对到参考基因组上,能比对上的比例为86.63%~88.59%(表5)。
表 5 比对核糖体后得到的未标记序列与参考基因组的比对统计Table 5. Statistics of unmapped reads and reference genome样品
Sample序列总数
Total reads/条唯一比对上的序列
Unique mapped reads多处比对上的序列
Multiple mapped reads数量
Number/条占比
Ratio/%数量
Number/条占比
Ratio/%CK-1 47756642 41591814 87.09 672956 1.41 CK-2 55033864 47903535 87.04 849190 1.54 CK-3 56270128 48813881 86.75 868458 1.54 T-1 53290572 46327128 86.93 743356 1.39 T-2 59953992 51103454 85.24 834008 1.39 T-3 51239894 44511502 86.87 705832 1.38 2.3 样本基因的注释和差异表达基因的筛选分析
利用Cufflinks进行转录本组装,利用Cuffmerge对多个样品的组装结果进行合并,共得到21 497个基因,其中20 573基因在参考基因组中得到注释,有924个新基因没有得到注释。将这些新基因的转录本在Nr、KEGG 数据库进行注释。
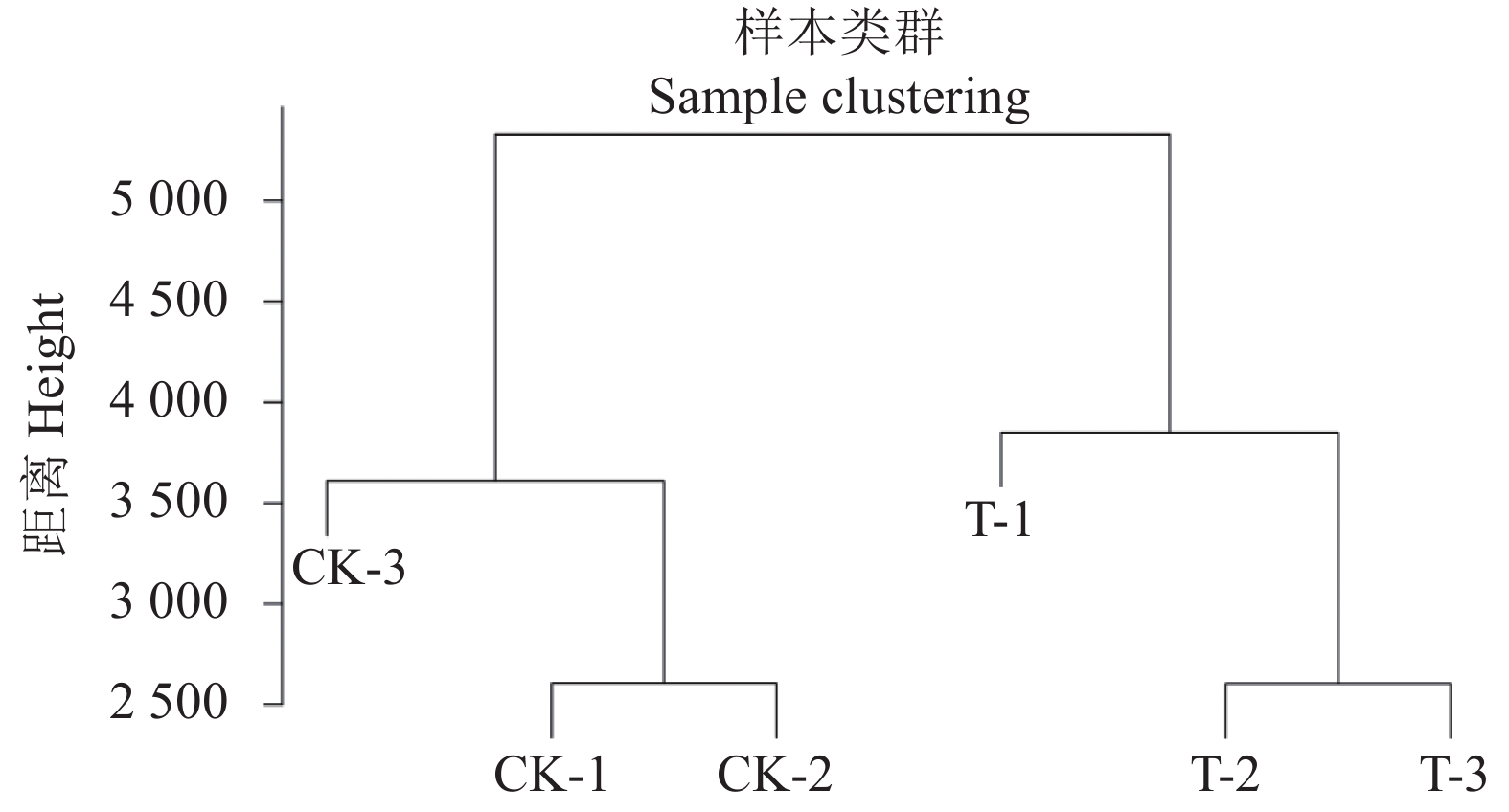
用全体基因的表达量对所有样本的关系进行层级聚类,层级聚类图可以真实地反映出每个样本与其他样本的关系,图2显示对照组3个重复聚为一类,处理组3个重复聚为一类,由此可以得知本试验处理重复性好。
利用 FDR 与 log2(FC) 来筛选差异基因,筛选条件为 FDR<0.05 且 |log2(FC)|>1,结果显示与对照相比,酸雨处理24 h后组中有879个基因显著上调,588个基因显著下调。筛选出样本中前50个DEGs(差异基因),均为上调表达基因,大部分涉及代谢途径、次生代谢、苯丙基类丙烷生物合成和萜类物质合成等相关基因。
2.4 差异表达基因的Gene Ontology富集分析
Gene Ontology(简称GO)是一个国际标准化的基因功能分类体系,分别从分子功能(Molecular function)、细胞组分(Cellular component)和生物过程(Biological process)来描述基因,能够全面描述生物体中基因和基因产物的属性,该体系随着研究的发展而动态更新。对筛选出的差异表达基因做GO功能分析,阐明处理差异在基因功能上的体现。在GO数据库中,细胞组成方面有2 101条基因,有97条显著差异基因,被注释到56条GO term中,其中GO:0005576 extracelluar region(细胞外区域)为差异表达基因中最显著富集的一个GO term,其次是GO:0030312 external encapsulating structure。在GO数据库中,分子功能方面有4 686条基因,有279条显著差异基因,被注释到206条GO term中,其中GO:0003824 catalytic activity(催化剂活性)为在差异表达基因中最显著富集的一个GO term,其次是GO:0016491 oxidoreductase activity(氧化还原酶活性)。在GO数据库中,生物过程方面有4 355条基因,有245条显著差异基因,被注释到409条GO term中,其中GO:0006259 DNA metabolic process(DNA代谢过程)为在差异表达基因中最显著富集的一个GO term,其次是GO:0009620 response to fungus(对真菌的响应)。表6是对差异基因按上下调进行GO term的分类统计。
表 6 差异基因GO功能分类Table 6. The Go functional classification of the differential expression本体
Ontology分类
Class上调表达
基因数量
Up-regulated
DEG number下调表达
基因数量
Down-regulated
DEG number本体
Ontology分类
Class上调表达
基因数量
Up-regulated
DEG number下调表达
基因数量
Down-regulated
DEG number生物过程
Biological
Process单一的生物过程
single-organism process90 45 分子功能
Molecular Function催化剂活性
catalytic activity163 81 细胞杀伤
cell killing1 0 转运因子活性
transporter activity19 3 刺激应答
response to stimulus24 7 信号转导过程
signal transducer activity5 2 细胞成分组织
cellular component organization or biogenesis8 12 分子功能调节
molecular function regulator1 1 免疫系统过程
immune system process2 0 核苷酸结合转录因子活性
nucleic acid binding transcription factor activity3 0 多组织过程/多机体过程
multi-organism process5 0 结合/结合剂活性
binding74 54 定位
localization29 5 结构分子活性
structural molecule activity1 0 再生
reproduction4 1 细胞成分
Cellular Component细胞间区域
extracellular region6 2 生长
growth0 1 细胞连丝
cell junction4 0 生物调控
biological regulation13 7 细胞膜要素
membrane part36 5 再生过程
reproductive process2 0 细胞膜 membrane 38 6 多细胞组织过程
multicellular organismal process1 2 膜结合腔体
membrane-enclosed lumen0 2 发育过程
developmental process2 2 细胞器
organelle11 23 细胞过程
cellular process78 64 细胞
cell23 26 信号
signaling2 2 细胞要素
cell part23 26 代谢过程
metabolic process116 68 大分子复合物
macromolecular complex3 8 细胞器要素
organelle part2 4 2.5 差异表达基因的KEGG富集分析
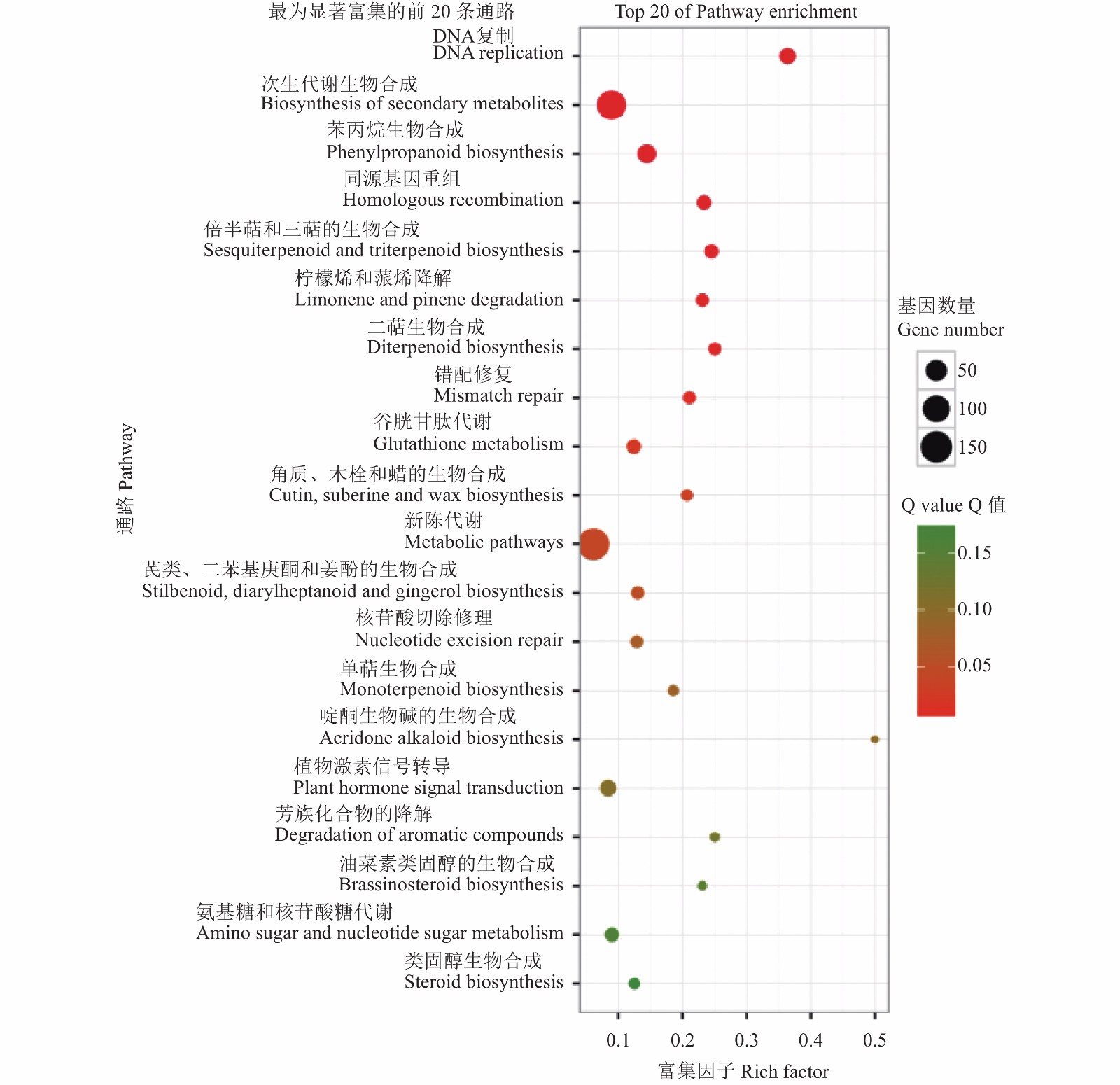
KEGG是代谢通路(Pathway)的主要公共数据库,差异表达基因Pathway显著性富集分析能确定这些差异表达基因参与的最主要的代谢和信号转导途径。本研究中有5 845条基因被注释到KEGG数据库的99条Pathway中,对照和处理组间有303条基因表达差异显著。图3显示差异表达基因中最为显著富集的前20条Pathway(按照Qvalue),其中ko03030 DNA replication Pathway 是差异表达基因中最显著富集的Pathway,在这条通路中有20个基因显著差异表达,与对照相比多为上调表达;其次是次生代谢的生物合成,在这条通路中有121个基因显著差异表达,与对照相比多为上调表达;再次是苯丙素类合成途径,有33个基因显著差异表达,与对照相比多为上调表达;接着是同源重组通路,有14个基因显著差异表达,与对照相比均为下调表达。
![]() 图 3 KEEP 代谢途径中最为显著富集的前20条通路注:Rich factor指差异表达的基因中位于该pathway条目的基因数目与所有基因中位于该pathway条目的基因总数的比值, Rich factor越大,表示富集的程度越高。Q value是做过多重假设检验校正之后的P value,取值范围为0到1,越接近于零,表示富集越显著。Figure 3. Top 20 enrichment pathways as analyzed by KEEPNote: Rich factor refers to ratio of number of DEGs located in pathway entry to number of all genes located in same pathway entry; high rich factor indicates high degree of enrichment. Q value is the P value after multiple hypothesis testing and corrections which ranges from 0 to 1 with 0 being most significantly enriched.
图 3 KEEP 代谢途径中最为显著富集的前20条通路注:Rich factor指差异表达的基因中位于该pathway条目的基因数目与所有基因中位于该pathway条目的基因总数的比值, Rich factor越大,表示富集的程度越高。Q value是做过多重假设检验校正之后的P value,取值范围为0到1,越接近于零,表示富集越显著。Figure 3. Top 20 enrichment pathways as analyzed by KEEPNote: Rich factor refers to ratio of number of DEGs located in pathway entry to number of all genes located in same pathway entry; high rich factor indicates high degree of enrichment. Q value is the P value after multiple hypothesis testing and corrections which ranges from 0 to 1 with 0 being most significantly enriched.2.6 次生代谢合成相关酶PCR分析
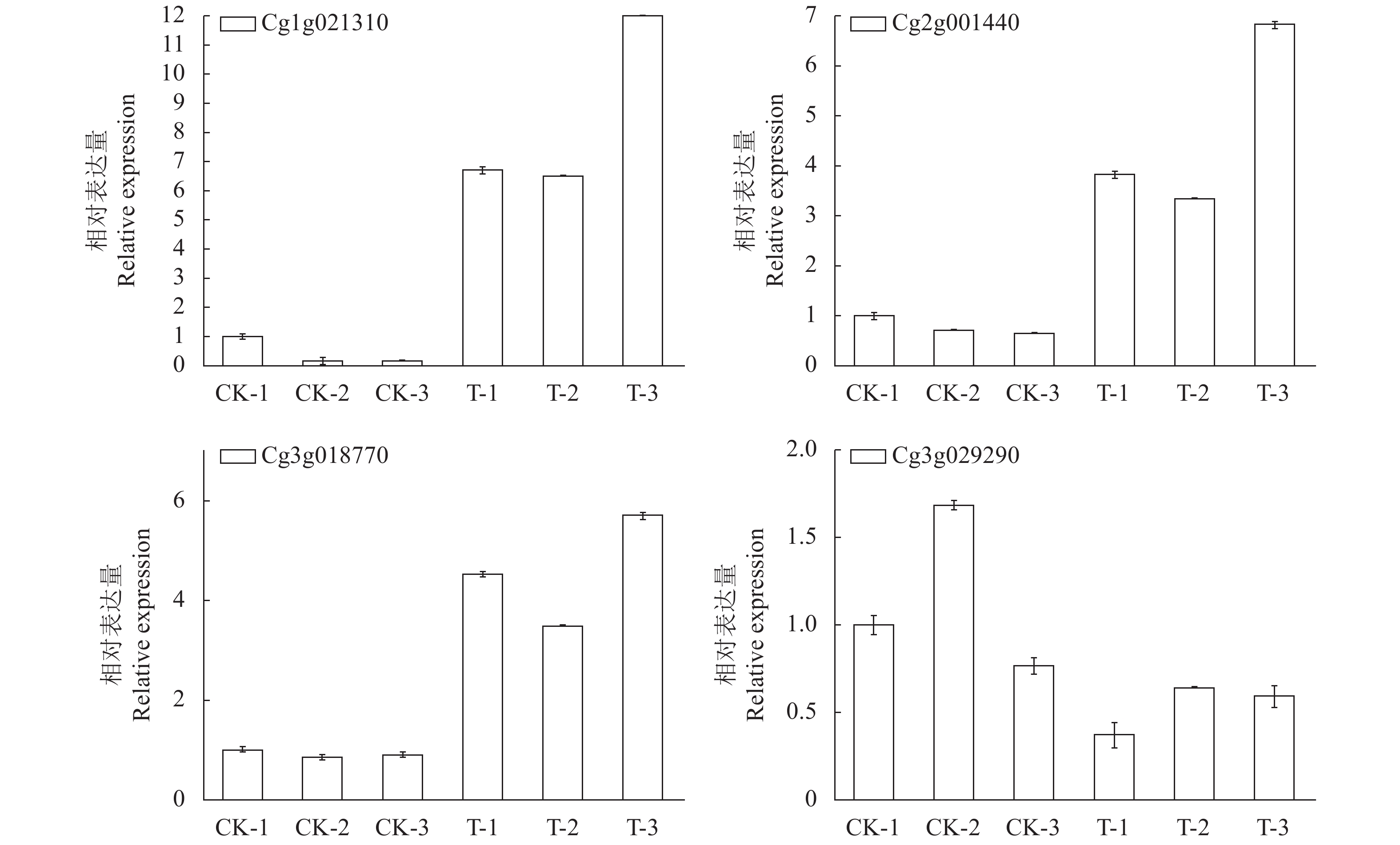
选择4个与次生代谢相关的差异显著基因[POD同工酶cg3g018770和cg2g001440、肉桂酰辅酶A还原酶(CCR)同工酶cg1g021310、4-香豆酸-辅酶a连接酶(4CL)同工酶cg3g029290]进行验证,经过验证发现这4个基因的实际情况与转录组数据一致,cg3g018770、cg2g001440和cg1g021310与对照相比均为上调表达,cg3g029290与对照相比明显下调,证明了转录组数据的可靠性(图4)。
3. 讨论与结论
酸雨对植物直接伤害表现为使叶子褪绿、黄化、出现块状伤斑、失水萎蔫和早落叶等症状,随着酸度增高和淋溶时间的延长受害越严重,一般嫩叶较老叶更易受到伤害[15]。叶片角质层是植物遭遇酸雨胁迫的第一道防线[15],琯溪蜜柚叶片具有较厚角质层,抗酸雨性较强,张琼等[12]研究表明中长期酸雨淋洗没有造成琯溪蜜柚叶片表面的可视性损害,本研究模拟酸雨淋洗的是较嫩叶片,叶片角质层较薄,24 h后就表现出较为明显的伤块,说明酸雨侵蚀嫩叶,产生了直接伤害。
酸雨通过气孔或被损害的叶片表皮进入细胞内部,会破坏代谢平衡,降低植物的光合作用[16-17],近些年来一些学者从分子生物学水平研究了酸雨对植物的影响。Hu等[6]研究认为酸雨胁迫可使得酸雨抗性物种大多参与基础代谢、细胞建成、光合和转录等过程的蛋白表达水平上调,使得酸雨敏感物种代谢、光合、信号转导和转录相关蛋白表达水平下调,本研究中酸雨处理24 h后琯溪蜜柚叶片有879个基因显著上调,588个基因显著下调,而筛选出样本中前50个DEGs(差异基因),均为上调表达基因,大部分涉及代谢途径、次生代谢、苯丙基类丙烷生物合成和萜类物质合成等相关基因,由此可以推测琯溪蜜柚对酸雨耐性较强。GO富集分析表明差异表达基因主要位于细胞外区域,催化剂活性是分子功能最为显著富集的GO term中,其次是氧化还原酶活性,生物过程中差异表达基因最显著的GO term是DNA 代谢过程。对差异基因对照KEGG数据库进行注释显示酸雨胁迫显著抑制DNA 复制通路,促进次生代谢的生物合成,尤其是苯丙素类合成途径。次生代谢是植物在长期进化中与环境相互作用的结果,是提高植物抗逆性的主要调节机制[18- 19]。由此可推测琯溪蜜柚主要是通过抑制DNA复制,影响细胞分裂增殖,进而抑制叶片生长,以此为代价使得植物将更多能量和资源分配给次生代谢来应对酸雨胁迫。选择次生代谢合成途径中3个主要基因[POD同工酶cg3g018770和cg2g001440、肉桂酰辅酶A还原酶(CCR)、同工酶cg1g021310]进行PCR荧光定量验证分析,结果也显示酸雨处理均明显促进了这3个基因的相对表达,进一步验证了转录组分析的结果。琯溪蜜柚通过调节次生代谢提高耐酸性与拟南芥耐酸机理相似,酸雨胁迫下拟南芥有多个与植物次生代谢相关基因表达量发生变化,木质素、生物碱等次生代谢产物的合成在酸雨处理下加强[11]。
本研究从表型和分子生物学角度研究酸雨胁迫对琯溪蜜柚的影响,结果表明pH 2.5的模拟酸雨侵蚀琯溪蜜柚嫩叶产生直接伤害;酸雨喷淋使得叶片部分基因表达差异显著,差异表达基因主要位于细胞外区域,对催化剂活性和氧化还原酶活性影响较大,酸雨胁迫显著抑制DNA 复制通路,促进次生代谢的生物合成,尤其是苯丙素类合成途径。由此可得出琯溪蜜柚对酸雨胁迫的响应是一个多基因参与、多个生物过程协同调控的过程,次生代谢的调节可能是应对酸雨胁迫的主要方式。
-
图 3 KEEP 代谢途径中最为显著富集的前20条通路
注:Rich factor指差异表达的基因中位于该pathway条目的基因数目与所有基因中位于该pathway条目的基因总数的比值, Rich factor越大,表示富集的程度越高。Q value是做过多重假设检验校正之后的P value,取值范围为0到1,越接近于零,表示富集越显著。
Figure 3. Top 20 enrichment pathways as analyzed by KEEP
Note: Rich factor refers to ratio of number of DEGs located in pathway entry to number of all genes located in same pathway entry; high rich factor indicates high degree of enrichment. Q value is the P value after multiple hypothesis testing and corrections which ranges from 0 to 1 with 0 being most significantly enriched.
表 1 qPCR 反应体系
Table 1 qPCR reaction system
qPCR反应体系
qPCR reaction system体积
Volume/μlqPCR反应混合物 qPCR Master Mix 10 PCR正向引物PCR Forward Primer/(10 mol·L−1) 0.4 PCR反向引物 PCR Reverse Primer/(10 mol·L−1) 0.4 cDNA模板 cDNA template 4 ddH2O 5.2 合计 Total 20 表 2 引物序列
Table 2 Primer sequence
基因
Gene序列
SequenceCg3g024100 F 5′CAATGTGAAGTCCAGCGTGTG3′ R 5′AGCCCTCGTAGTTCTCGTCA3′ Cg3g018770 F 5′CATTTACCAATCGCCTCTATCC3′ R 5′ACCCCTGTCGGTTCATCAAGT3′ Cg2g001440 F 5′GGCAATCCAGACCCAACACT3′ R 5′AATGGCAGCGGTATCGGC3′ Cg1g021310 F 5′GTTGACGAACATTGCTGGAGT3′ R 5′GCAGCAATGTCCCTATCACC3′ Cg3g029290 F 5′CGAAGTAGAGTCCCTCAAGCA3′ R 5′TGTCACCAGTATGAAGCCAACC3′ 表 3 有效数据评估统计
Table 3 Statistics of valid data
样本
Sample处理后
序列数据
Clean
Data/bp过滤后的
序列数据
HQ Clean
Data/bp处理后
序列数
Clean Reads
Num/条过滤后的序列
HQ Clean ReadsQ20 Q30 GC 数量
Number/条占比
Ratio/%数量
Number/bp占比
Ratio/%数量
Number/bp占比
Ratio/%数量
Number/bp占比
Ratio/ %CK-1 7361352600 7111541211 49075684 48127600 98.07 6999438082 98.42 6769749153 95.19 3153041858 44.34 CK-2 8465603100 8202153477 56437354 55469140 98.28 8080142163 98.51 7826038154 95.41 3620789302 44.14 CK-3 8683275600 8408343801 57888504 56867832 98.24 8281909562 98.5 8019114646 95.37 3726434942 44.32 T-1 8203212900 7937070962 54688086 53695546 98.19 7816272312 98.48 7566409577 95.33 3522622180 44.38 T-2 9249897000 8926570988 61665980 60549386 98.19 8792579583 98.5 8514792020 95.39 3962645890 44.39 T-3 7880490300 7621577079 52536602 51567848 98.16 7503238757 98.45 7259492038 95.25 3369836120 44.21 注:1.Q20-碱基质量 ≥20的碱基;2.Q30-碱基质量 ≥30的碱基;3.GC-鸟嘌呤和胞嘧啶;4.CK-1、CK-2、CK-3为清水对照3个重复,T-1、T-2、T-3为pH2.5模拟酸雨喷淋的3个重复,表4、5同,图2、4同。
Note: 1. Q20- Base quality≥20. 2. Q30- Base quality≥30.3. GC- Guanine and Cytosine.4.CK-1, ck-2 and ck-3 are three repetitions of clear water control, and T-1, T-2 and T-3 are three repetitions of pH2.5 simulated acid rain spray.The same as Tab. 4 and. The same as Fig.2 and 4.表 4 过滤后的序列与核糖体RNA 的比对统计
Table 4 Statistics of HQ-clean data and rRNA
样品
Sample过滤后的序列数
HQ Clean Reads
Number/条比对上rRNA的序列
Mapped Reads未比对上rRNA的序列
Unmapped Reads数量
Number/条占比
Ratio/%数量
Number/条占比
Ratio/%CK-1 48127600 370958 0.77 47756642 99.23 CK-2 55469140 435276 0.78 55033864 99.22 CK-3 56867832 597704 1.05 56270128 98.95 T-1 53695546 404974 0.75 53290572 99.25 T-2 60549386 595394 0.98 59953992 99.02 T-3 51567848 327954 0.64 51239894 99.36 表 5 比对核糖体后得到的未标记序列与参考基因组的比对统计
Table 5 Statistics of unmapped reads and reference genome
样品
Sample序列总数
Total reads/条唯一比对上的序列
Unique mapped reads多处比对上的序列
Multiple mapped reads数量
Number/条占比
Ratio/%数量
Number/条占比
Ratio/%CK-1 47756642 41591814 87.09 672956 1.41 CK-2 55033864 47903535 87.04 849190 1.54 CK-3 56270128 48813881 86.75 868458 1.54 T-1 53290572 46327128 86.93 743356 1.39 T-2 59953992 51103454 85.24 834008 1.39 T-3 51239894 44511502 86.87 705832 1.38 表 6 差异基因GO功能分类
Table 6 The Go functional classification of the differential expression
本体
Ontology分类
Class上调表达
基因数量
Up-regulated
DEG number下调表达
基因数量
Down-regulated
DEG number本体
Ontology分类
Class上调表达
基因数量
Up-regulated
DEG number下调表达
基因数量
Down-regulated
DEG number生物过程
Biological
Process单一的生物过程
single-organism process90 45 分子功能
Molecular Function催化剂活性
catalytic activity163 81 细胞杀伤
cell killing1 0 转运因子活性
transporter activity19 3 刺激应答
response to stimulus24 7 信号转导过程
signal transducer activity5 2 细胞成分组织
cellular component organization or biogenesis8 12 分子功能调节
molecular function regulator1 1 免疫系统过程
immune system process2 0 核苷酸结合转录因子活性
nucleic acid binding transcription factor activity3 0 多组织过程/多机体过程
multi-organism process5 0 结合/结合剂活性
binding74 54 定位
localization29 5 结构分子活性
structural molecule activity1 0 再生
reproduction4 1 细胞成分
Cellular Component细胞间区域
extracellular region6 2 生长
growth0 1 细胞连丝
cell junction4 0 生物调控
biological regulation13 7 细胞膜要素
membrane part36 5 再生过程
reproductive process2 0 细胞膜 membrane 38 6 多细胞组织过程
multicellular organismal process1 2 膜结合腔体
membrane-enclosed lumen0 2 发育过程
developmental process2 2 细胞器
organelle11 23 细胞过程
cellular process78 64 细胞
cell23 26 信号
signaling2 2 细胞要素
cell part23 26 代谢过程
metabolic process116 68 大分子复合物
macromolecular complex3 8 细胞器要素
organelle part2 4 -
[1] 林燕金, 林旗华, 姜翠翠, 等. 福建省柚类产业发展现状及对策 [J]. 东南园艺, 2014, 2(5):39−42. DOI: 10.3969/j.issn.1004-6089.2014.05.009 LIN Y J, LIN Q H, JIANG C C, et al. Industry status and countermeasures of pomelo industry in Fujian Province [J]. Southeast Horticulture, 2014, 2(5): 39−42.(in Chinese) DOI: 10.3969/j.issn.1004-6089.2014.05.009
[2] 王梨嬛, 潘永娟, 杨莉, 等. ‘琯溪蜜柚’荧光定量PCR内参基因的筛选 [J]. 果树学报, 2013, 30(1):48−54. WANG L H, PAN Y J, YANG L, et al. Validation of internal reference genes for qRT-PCR normalization in Guanxi Sweet Pummelo(Citrus grandis) [J]. Journal of Fruit Science, 2013, 30(1): 48−54.(in Chinese)
[3] 赵卫红. 福建省主要城市降水离子特征及沉降量现状分析 [J]. 亚热带资源与环境学报, 2008, 3(3):19−24. DOI: 10.3969/j.issn.1673-7105.2008.03.004 ZHAO W H. On characteristics of precipitation ion and current sedimentation situation in main cities of Fujian Province [J]. Journal of Subtropical Resources and Environment, 2008, 3(3): 19−24.(in Chinese) DOI: 10.3969/j.issn.1673-7105.2008.03.004
[4] 钱笑杰, 林晓兰, 肖靖, 等. 福建果园土壤pH值、养分关系与土壤肥力质量评价研究: 以福建省漳州市平和县琯溪蜜柚园地为例 [J]. 福建热作科技, 2017, 42(1):9−15. DOI: 10.3969/j.issn.1006-2327.2017.01.003 QIAN X J, LIN X L, XIAO J, et al. Evaluation research between soil pH, nutrition and soil fertility in Fujian orchard [J]. Fujian Science & Technology of Tropical Crops, 2017, 42(1): 9−15.(in Chinese) DOI: 10.3969/j.issn.1006-2327.2017.01.003
[5] 任晓巧, 章家恩, 向慧敏, 等. 酸雨对植物地上部生理生态的影响研究进展与展望 [J]. 应用与环境生物学报, 2021, 27(5):1−10. REN X Q, ZHANG J E, XIANG H M, et al. Research advances and prospects for effects of acid rain on the aboveground physiology of plants and the related alleviation countermeasures [J]. Chinese Journal of Applied & Environmental Biology, 2021, 27(5): 1−10.(in Chinese)
[6] HU W J, CHEN J, LIU T W, et al. Proteome and calcium-related gene expression in Pinus massoniana needles in response to acid rain under different calcium levels [J]. Plant and Soil, 2014, 380: 285−303. DOI: 10.1007/s11104-014-2086-9
[7] 宋晓梅, 曹向阳. 模拟酸雨对不同园林植物叶片生理生态特性的影响 [J]. 水土保持研究, 2017, 24(2):365−370. SONG X M, CAO X Y. Effect of simulated acid rain on the physiological and ecological characteristics of different garden plants [J]. Research of Soil and Water Conservation, 2017, 24(2): 365−370.(in Chinese)
[8] YAN P, WU L Q, WANG D H, et al. Soil acidification in Chinese tea plantations [J]. Science of the Total Environment, 2020, 715: 1−7.
[9] LIU T W, NIU L, FU B, et al. A transcriptomic study reveals differentially expressed genes and pathways respond to simulated acid rain in Arabidopsis thaliana [J]. Genome, 2013, 56(1): 49−60. DOI: 10.1139/gen-2012-0090
[10] 俞翰炳. 模拟酸雨对拟南芥抗病性的影响及机理探究[D]. 杭州: 杭州师范大学, 2015. YU H B. Studies on effects and mechanisms of simulated acid rain on Arabidopsis thaliana tolerance to disease[D]. Hangzhou: Hangzhou Normal University, 2015. (in Chinese).
[11] 牛力. 模拟酸雨对拟南芥某些生理特性和基因表达谱的影响[D]. 厦门: 厦门大学, 2009. NIU L. Effects of simulated acid rain on some physiological characteristics and gene expression profiles of Arabidopsis thaliana[D]. Xiamen, China: Xiamen University, 2009. (in Chinese).
[12] 张琼, 陆銮眉, 戴清霞, 等. 模拟酸雨对‘琯溪蜜柚’叶片抗氧化酶活性和光合作用的影响 [J]. 果树学报, 2018, 35(7):828−835. ZHANG Q, LU L M, DAI Q X, et al. Effects of acid rain on the leaves antioxidase activity and photosynthesis of Citrus Grandis(L.) Osbeck. ‘Guanximiyou’ seedlings [J]. Journal of Fruit Science, 2018, 35(7): 828−835.(in Chinese)
[13] KIM D, PERTEA G, TRAPNELL C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions [J]. Genome Biology, 2013, 14(4): R36. DOI: 10.1186/gb-2013-14-4-r36
[14] TRAPNELL C, WILLIAMS B A, PERTEA G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation [J]. Nature Biotechnology, 2010, 28(5): 511−515. DOI: 10.1038/nbt.1621
[15] 任晓巧, 章家恩, 向慧敏, 等. 酸雨对植物地上部生理生态的影响研究进展与展望[J/OL]. 应用与环境生物学报: 1-12 [2021-04-25]. https://doi.org/10.19675/j.cnki.1006-687x.2020.07054. REN X Q, ZHANG J, XIANG H M, et al. Research advances and prospects for effects of acid rain on the aboveground physiology of plants and the related alleviation countermeasures [J/OL]. Chinese Journal of Applied and Environmental Biology: 1−12 [2021-04-25]. https://doi.org/10.19675/j.cnki.1006-687x.2020.07054. (in Chinese).
[16] 张利霞, 郝国宝, 常青山, 等. 模拟酸雨胁迫对夏枯草抗氧化酶活性和光合参数的影响 [J]. 中国草地学报, 2020, 42(4):56−61. ZHANG L X, HAO G B, CHANG Q S, et al. Antioxidant capacity and photosynthetic characteristics of Prunella vulgaris seedlings in response to simulated acid rain stress [J]. Chinese Journal of Grassland, 2020, 42(4): 56−61.(in Chinese)
[17] 马永佳, 梁婵娟. 外源钙对模拟酸雨胁迫下水稻质膜组分和钙形态的调节 [J]. 农业环境科学学报, 2021, 40(6): 1159-1166. MA Y J, LIANG C J. Regulation of exogenous calcium on plasma membrane compositions and calcium forms of rice roots under simulated acid rain stress [J]. Journal of Agro-Environment Science, 2021, 40(6): 1159−1166. https://kns.cnki.net/kcms/detail/12.1347.
[18] 刘洪博, 刘新龙, 苏火生, 等. 干旱胁迫下割手密根系转录组差异表达分析 [J]. 中国农业科学, 2017, 50(6):1167−1178. DOI: 10.3864/j.issn.0578-1752.2017.06.017 LIU H B, LIU X L, SU H S, et al. Transcriptome difference analysis of Saccharum spontaneum roots in response to drought stress [J]. Scientia Agricultura Sinica, 2017, 50(6): 1167−1178.(in Chinese) DOI: 10.3864/j.issn.0578-1752.2017.06.017
[19] 阎秀峰. 植物次生代谢生态学 [J]. 植物生态学报, 2001, 25(5):639−640, 622. DOI: 10.3321/j.issn:1005-264X.2001.05.021 YAN X F. Ecology of plant secondary metabolism [J]. Acta Phytoecologica Sinica, 2001, 25(5): 639−640, 622.(in Chinese) DOI: 10.3321/j.issn:1005-264X.2001.05.021
-
期刊类型引用(1)
1. 张琼,陆銮眉,胡元庆,朱丽霞,李媛媛,郑思淼. 土壤pH对琯溪蜜柚叶片枝条和果实的影响. 生态学杂志. 2024(05): 1291-1299 .  百度学术
百度学术
其他类型引用(0)




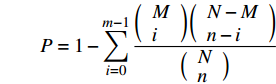
 下载:
下载: